Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations
- PMID: 21757228
- PMCID: PMC3155290
- DOI: 10.1016/j.cell.2011.06.019
Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations
Erratum in
- Cell. 2011 Aug 19;146(4):659
Abstract
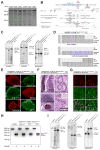
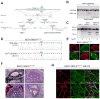
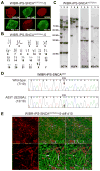
Patient-specific induced pluripotent stem cells (iPSCs) derived from somatic cells provide a unique tool for the study of human disease, as well as a promising source for cell replacement therapies. One crucial limitation has been the inability to perform experiments under genetically defined conditions. This is particularly relevant for late age onset disorders in which in vitro phenotypes are predicted to be subtle and susceptible to significant effects of genetic background variations. By combining zinc finger nuclease (ZFN)-mediated genome editing and iPSC technology, we provide a generally applicable solution to this problem, generating sets of isogenic disease and control human pluripotent stem cells that differ exclusively at either of two susceptibility variants for Parkinson's disease by modifying the underlying point mutations in the α-synuclein gene. The robust capability to genetically correct disease-causing point mutations in patient-derived hiPSCs represents significant progress for basic biomedical research and an advance toward hiPSC-based cell replacement therapies.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Gene editing in stem cells hits the target.Cell Stem Cell. 2011 Aug 5;9(2):93-4. doi: 10.1016/j.stem.2011.07.011. Cell Stem Cell. 2011. PMID: 21816359
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

