Global profiling of DNA methylation erasure in mouse primordial germ cells
- PMID: 22357612
- PMCID: PMC3317146
- DOI: 10.1101/gr.130997.111
Global profiling of DNA methylation erasure in mouse primordial germ cells
Abstract
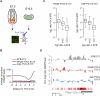
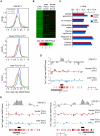
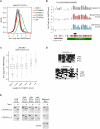
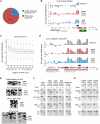
Epigenetic reprogramming, characterized by loss of cytosine methylation and histone modifications, occurs during mammalian development in primordial germ cells (PGCs), yet the targets and kinetics of this process are poorly characterized. Here we provide a map of cytosine methylation on a large portion of the genome in developing male and female PGCs isolated from mouse embryos. We show that DNA methylation erasure is global and affects genes of various biological functions. We also reveal complex kinetics of demethylation that are initiated at most genes in early PGC precursors around embryonic day 8.0-9.0. In addition, besides intracisternal A-particles (IAPs), we identify rare LTR-ERV1 retroelements and single-copy sequences that resist global methylation erasure in PGCs as well as in preimplantation embryos. Our data provide important insights into the targets and dynamics of DNA methylation reprogramming in mammalian germ cells.
Figures

References
-
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42: 1093–1100 - PubMed
-
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424 - PubMed
-
- Guibert S, Forne T, Weber M 2009. Dynamic regulation of DNA methylation during mammalian development. Epigenomics 1: 81–98 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
