Whole-genome association study on tissue tropism phenotypes in group A Streptococcus
- PMID: 21949075
- PMCID: PMC3232895
- DOI: 10.1128/JB.05263-11
Whole-genome association study on tissue tropism phenotypes in group A Streptococcus
Abstract
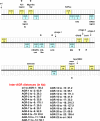
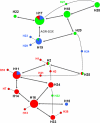
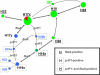
Group A Streptococcus (GAS) has a rich evolutionary history of horizontal transfer among its core genes. Yet, despite extensive genetic mixing, GAS strains have discrete ecological phenotypes. To further our understanding of the molecular basis for ecological phenotypes, comparative genomic hybridization of a set of 97 diverse strains to a GAS pangenome microarray was undertaken, and the association of accessory genes with emm genotypes that define tissue tropisms for infection was determined. Of the 22 nonprophage accessory gene regions (AGRs) identified, only 3 account for all statistically significant linkage disequilibrium among strains having the genotypic biomarkers for throat versus skin infection specialists. Networked evolution and population structure analyses of loci representing each of the AGRs reveal that most strains with the skin specialist and generalist biomarkers form discrete clusters, whereas strains with the throat specialist biomarker are highly diverse. To identify coinherited and coselected accessory genes, the strength of genetic associations was determined for all possible pairwise combinations of accessory genes among the 97 GAS strains. Accessory genes showing very strong associations provide the basis for an evolutionary model, which reveals that a major transition between many throat and skin specialist haplotypes correlates with the gain or loss of genes encoding fibronectin-binding proteins. This study employs a novel synthesis of tools to help delineate the major genetic changes associated with key adaptive shifts in an extensively recombined bacterial species.
Figures




References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Anthony B. F., Kaplan E. L., Wannamaker L. W., Chapman S. S. 1976. The dynamics of streptococcal infections in a defined population of children: serotypes associated with skin and respiratory infections. Am. J. Epidemiol. 104:652–666 - PubMed
-
- Banks D. J., Porcella S. F., Barbian K. D., Martin J. M., Musser J. M. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J. Infect. Dis. 188:1898–1908 - PubMed
-
- Beall B. 16 May 2011, posting date Streptococcus pyogenes emm sequence database. http://cdc.gov/ncidod/biotech/strep/strepblast.htm
-
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

