Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos
- PMID: 22266940
- PMCID: PMC3477627
- DOI: 10.1038/nature10756
Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos
Abstract
In animals, maternal gene products deposited into eggs regulate embryonic development before activation of the zygotic genome. In plants, an analogous period of prolonged maternal control over embryogenesis is thought to occur based on some gene-expression studies. However, other gene-expression studies and genetic analyses show that some transcripts must derive from the early zygotic genome, implying that the prevailing model does not fully explain the nature of zygotic genome activation in plants. To determine the maternal, paternal and zygotic contributions to the early embryonic transcriptome, we sequenced the transcripts of hybrid embryos from crosses between two polymorphic inbred lines of Arabidopsis thaliana and used single-nucleotide polymorphisms diagnostic of each parental line to quantify parental contributions. Although some transcripts seemed to be either inherited from primarily one parent or transcribed from imprinted loci, the vast majority of transcripts were produced in near-equal amounts from both maternal and paternal alleles, even during the initial stages of embryogenesis. Results of reporter experiments and analyses of transcripts from genes that are not expressed in sperm and egg indicate early and widespread zygotic transcription. Thus, in contrast to early animal embryogenesis, early plant embryogenesis is mostly under zygotic control.
Figures
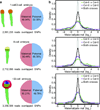
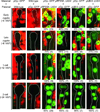
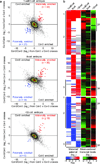
Comment in
-
Plant biology: Equal-parenting policy.Nature. 2012 Jan 22;482(7383):42-4. doi: 10.1038/nature10869. Nature. 2012. PMID: 22266937 No abstract available.
-
Development: Parental influences on plant development.Nat Rev Genet. 2012 Jan 31;13(3):147. doi: 10.1038/nrg3175. Nat Rev Genet. 2012. PMID: 22290415 No abstract available.
References
-
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. - PubMed
-
- Autran D, et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145(5):707–719. - PubMed
-
- Vielle-Calzada JP, Baskar R, Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404(6773):91–94. - PubMed
-
- Baroux C, Blanvillain R, Gallois P. Paternally inherited transgenes are down-regulated but retain low activity during early embryogenesis in Arabidopsis. FEBS Lett. 2001;509(1):11–16. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

