Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon
- PMID: 22730287
- PMCID: PMC3458547
- DOI: 10.1093/nar/gks594
Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon
Abstract
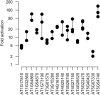
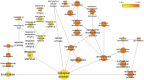
The plant-specific, B3 domain-containing transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) is an essential component of the regulatory network controlling the development and maturation of the Arabidopsis thaliana seed. Genome-wide chromatin immunoprecipitation (ChIP-chip), transcriptome analysis, quantitative reverse transcriptase-polymerase chain reaction and a transient promoter activation assay have been combined to identify a set of 98 ABI3 target genes. Most of these presumptive ABI3 targets require the presence of abscisic acid for their activation and are specifically expressed during seed maturation. ABI3 target promoters are enriched for G-box-like and RY-like elements. The general occurrence of these cis motifs in non-ABI3 target promoters suggests the existence of as yet unidentified regulatory signals, some of which may be associated with epigenetic control. Several members of the ABI3 regulon are also regulated by other transcription factors, including the seed-specific, B3 domain-containing FUS3 and LEC2. The data strengthen and extend the notion that ABI3 is essential for the protection of embryonic structures from desiccation and raise pertinent questions regarding the specificity of promoter recognition.
Figures




References
-
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12:294–300. - PubMed
-
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. - PubMed
-
- Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 2005;49:645–651. - PubMed
-
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

