Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression
- PMID: 22406644
- PMCID: PMC3378637
- DOI: 10.1038/ng.2204
Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression
Abstract
Although genome-wide association studies (GWAS) have identified the existence of numerous population-based cancer susceptibility loci, mechanistic insights remain limited, particularly for intergenic polymorphisms. Here, we show that polymorphism at a remote intergenic region on chromosome 11q13.3, recently identified as a susceptibility locus for renal cell carcinoma, modulates the binding and function of hypoxia-inducible factor (HIF) at a previously unrecognized transcriptional enhancer of CCND1 (encoding cyclin D1) that is specific for renal cancers characterized by inactivation of the von Hippel-Lindau tumor suppressor (pVHL). The protective haplotype impairs binding of HIF-2, resulting in an allelic imbalance in cyclin D1 expression, thus affecting a link between hypoxia pathways and cell cycle control.
Figures
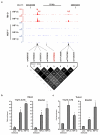
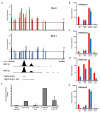
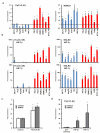
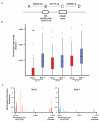

Comment in
-
Cancer genetics: HIF enhances its reputation.Nat Rev Cancer. 2012 Apr 12;12(5):316. doi: 10.1038/nrc3266. Nat Rev Cancer. 2012. PMID: 22495323 No abstract available.
References
-
- Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. - PubMed
-
- Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. - PubMed
-
- Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. - PubMed
References for Methods section
-
- Brown J, et al. Subtelomeric chromosome rearrangements are detected using an innovative 12-color FISH assay (M-TEL) Nat Med. 2001;7:497–501. - PubMed
-
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42.
-
- Hagege H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–33. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

