Genome features of "Dark-fly", a Drosophila line reared long-term in a dark environment
- PMID: 22432011
- PMCID: PMC3303825
- DOI: 10.1371/journal.pone.0033288
Genome features of "Dark-fly", a Drosophila line reared long-term in a dark environment
Abstract
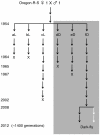
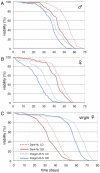
Organisms are remarkably adapted to diverse environments by specialized metabolisms, morphology, or behaviors. To address the molecular mechanisms underlying environmental adaptation, we have utilized a Drosophila melanogaster line, termed "Dark-fly", which has been maintained in constant dark conditions for 57 years (1400 generations). We found that Dark-fly exhibited higher fecundity in dark than in light conditions, indicating that Dark-fly possesses some traits advantageous in darkness. Using next-generation sequencing technology, we determined the whole genome sequence of Dark-fly and identified approximately 220,000 single nucleotide polymorphisms (SNPs) and 4,700 insertions or deletions (InDels) in the Dark-fly genome compared to the genome of the Oregon-R-S strain, a control strain. 1.8% of SNPs were classified as non-synonymous SNPs (nsSNPs: i.e., they alter the amino acid sequence of gene products). Among them, we detected 28 nonsense mutations (i.e., they produce a stop codon in the protein sequence) in the Dark-fly genome. These included genes encoding an olfactory receptor and a light receptor. We also searched runs of homozygosity (ROH) regions as putative regions selected during the population history, and found 21 ROH regions in the Dark-fly genome. We identified 241 genes carrying nsSNPs or InDels in the ROH regions. These include a cluster of alpha-esterase genes that are involved in detoxification processes. Furthermore, analysis of structural variants in the Dark-fly genome showed the deletion of a gene related to fatty acid metabolism. Our results revealed unique features of the Dark-fly genome and provided a list of potential candidate genes involved in environmental adaptation.
Conflict of interest statement
Figures



References
-
- Nadeau NJ, Jiggins CD. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 2010;26:484–492. - PubMed
-
- Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. - PubMed
-
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. - PubMed
-
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

