Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function
- PMID: 22520852
- PMCID: PMC3551341
- DOI: 10.1016/j.immuni.2012.02.017
Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function
Abstract
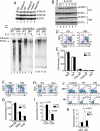
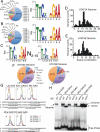
Cytokine-activated STAT proteins dimerize and bind to high-affinity motifs, and N-terminal domain-mediated oligomerization of dimers allows tetramer formation and binding to low-affinity tandem motifs, but the functions of dimers versus tetramers are unknown. We generated Stat5a-Stat5b double knockin (DKI) N-domain mutant mice in which STAT5 proteins form dimers but not tetramers, identified cytokine-regulated genes whose expression required STAT5 tetramers, and defined dimer versus tetramer consensus motifs. Whereas Stat5-deficient mice exhibited perinatal lethality, DKI mice were viable; thus, STAT5 dimers were sufficient for survival. Nevertheless, STAT5 DKI mice had fewer CD4(+)CD25(+) T cells, NK cells, and CD8(+) T cells, with impaired cytokine-induced and homeostatic proliferation of CD8(+) T cells. Moreover, DKI CD8(+) T cell proliferation after viral infection was diminished and DKI Treg cells did not efficiently control colitis. Thus, tetramerization of STAT5 is critical for cytokine responses and normal immune function, establishing a critical role for STAT5 tetramerization in vivo.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





Comment in
-
STATus report on tetramers.Immunity. 2012 Apr 20;36(4):553-5. doi: 10.1016/j.immuni.2012.04.003. Immunity. 2012. PMID: 22520849
References
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
-
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. - PubMed
-
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

