Xenobiotics shape the physiology and gene expression of the active human gut microbiome
- PMID: 23332745
- PMCID: PMC3552296
- DOI: 10.1016/j.cell.2012.10.052
Xenobiotics shape the physiology and gene expression of the active human gut microbiome
Abstract
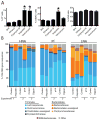
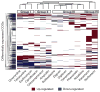
The human gut contains trillions of microorganisms that influence our health by metabolizing xenobiotics, including host-targeted drugs and antibiotics. Recent efforts have characterized the diversity of this host-associated community, but it remains unclear which microorganisms are active and what perturbations influence this activity. Here, we combine flow cytometry, 16S rRNA gene sequencing, and metatranscriptomics to demonstrate that the gut contains a distinctive set of active microorganisms, primarily Firmicutes. Short-term exposure to a panel of xenobiotics significantly affected the physiology, structure, and gene expression of this active gut microbiome. Xenobiotic-responsive genes were found across multiple bacterial phyla, encoding antibiotic resistance, drug metabolism, and stress response pathways. These results demonstrate the power of moving beyond surveys of microbial diversity to better understand metabolic activity, highlight the unintended consequences of xenobiotics, and suggest that attempts at personalized medicine should consider interindividual variations in the active human gut microbiome.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Dyeing to learn more about the gut microbiota.Cell Host Microbe. 2013 Feb 13;13(2):119-20. doi: 10.1016/j.chom.2013.01.011. Cell Host Microbe. 2013. PMID: 23414750 Free PMC article.
-
Xenobiotics and the human gut microbiome: metatranscriptomics reveal the active players.Cell Metab. 2013 Mar 5;17(3):317-8. doi: 10.1016/j.cmet.2013.02.013. Cell Metab. 2013. PMID: 23473028 Free PMC article.
References
-
- Baselt RC. Disposition of toxic drugs and chemicals in man. 8. Foster City, California: Biomedical Publications; 2008.
-
- Blaut M. Ecology and physiology of the intestinal tract. In: Compans RW, Cooper MD, Honjo T, Koprowski H, Melchers F, Oldstone MBA, Vogt PK, Gleba YY, Malissen B, Aktories K, editors. Curr Top Microbiol Immunol. Berlin Heidelberg: Springer-Verlag; 2011.
-
- Bouvier T, del Giorgio PA, Gasol JM. A comparative study of the cytometric characteristics of High and Low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ Microbiol. 2007;9:2050–2066. - PubMed
-
- Bouvier T, Maurice CF. A single-cell analysis of virioplankton adsorption, infection, and intracellular abundance in different bacterioplankton physiologic categories. Microb Ecol. 2011;62:669–678. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

