Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity
- PMID: 23103910
- PMCID: PMC3507454
- DOI: 10.1038/ncb2603
Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity
Abstract
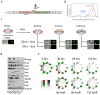
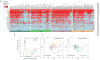
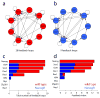
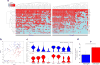
A number of key regulators of mouse embryonic stem (ES) cell identity, including the transcription factor Nanog, show strong expression fluctuations at the single-cell level. The molecular basis for these fluctuations is unknown. Here we used a genetic complementation strategy to investigate expression changes during transient periods of Nanog downregulation. Employing an integrated approach that includes high-throughput single-cell transcriptional profiling and mathematical modelling, we found that early molecular changes subsequent to Nanog loss are stochastic and reversible. However, analysis also revealed that Nanog loss severely compromises the self-sustaining feedback structure of the ES cell regulatory network. Consequently, these nascent changes soon become consolidated to committed fate decisions in the prolonged absence of Nanog. Consistent with this, we found that exogenous regulation of Nanog-dependent feedback control mechanisms produced a more homogeneous ES cell population. Taken together our results indicate that Nanog-dependent feedback loops have a role in controlling both ES cell fate decisions and population variability.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. - PubMed
-
- Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. - PubMed
-
- Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–U1238. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

