Comparative analysis of RNA sequencing methods for degraded or low-input samples
- PMID: 23685885
- PMCID: PMC3821180
- DOI: 10.1038/nmeth.2483
Comparative analysis of RNA sequencing methods for degraded or low-input samples
Erratum in
- Nat Methods. 2014 Feb;11(2):210
Abstract
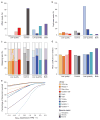
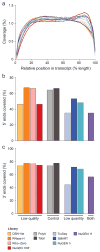
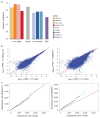
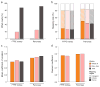
RNA-seq is an effective method for studying the transcriptome, but it can be difficult to apply to scarce or degraded RNA from fixed clinical samples, rare cell populations or cadavers. Recent studies have proposed several methods for RNA-seq of low-quality and/or low-quantity samples, but the relative merits of these methods have not been systematically analyzed. Here we compare five such methods using metrics relevant to transcriptome annotation, transcript discovery and gene expression. Using a single human RNA sample, we constructed and sequenced ten libraries with these methods and compared them against two control libraries. We found that the RNase H method performed best for chemically fragmented, low-quality RNA, and we confirmed this through analysis of actual degraded samples. RNase H can even effectively replace oligo(dT)-based methods for standard RNA-seq. SMART and NuGEN had distinct strengths for measuring low-quantity RNA. Our analysis allows biologists to select the most suitable methods and provides a benchmark for future method development.
Figures


References
-
- Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

