Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells
- PMID: 23685454
- PMCID: PMC3683364
- DOI: 10.1038/nature12172
Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells
Abstract
Recent molecular studies have shown that, even when derived from a seemingly homogenous population, individual cells can exhibit substantial differences in gene expression, protein levels and phenotypic output, with important functional consequences. Existing studies of cellular heterogeneity, however, have typically measured only a few pre-selected RNAs or proteins simultaneously, because genomic profiling methods could not be applied to single cells until very recently. Here we use single-cell RNA sequencing to investigate heterogeneity in the response of mouse bone-marrow-derived dendritic cells (BMDCs) to lipopolysaccharide. We find extensive, and previously unobserved, bimodal variation in messenger RNA abundance and splicing patterns, which we validate by RNA-fluorescence in situ hybridization for select transcripts. In particular, hundreds of key immune genes are bimodally expressed across cells, surprisingly even for genes that are very highly expressed at the population average. Moreover, splicing patterns demonstrate previously unobserved levels of heterogeneity between cells. Some of the observed bimodality can be attributed to closely related, yet distinct, known maturity states of BMDCs; other portions reflect differences in the usage of key regulatory circuits. For example, we identify a module of 137 highly variable, yet co-regulated, antiviral response genes. Using cells from knockout mice, we show that variability in this module may be propagated through an interferon feedback circuit, involving the transcriptional regulators Stat2 and Irf7. Our study demonstrates the power and promise of single-cell genomics in uncovering functional diversity between cells and in deciphering cell states and circuits.
Figures
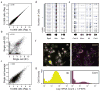

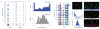

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- F32 HD075541/HD/NICHD NIH HHS/United States
- 1F32HD075541-01/HD/NICHD NIH HHS/United States
- 5DP1OD003893-03/OD/NIH HHS/United States
- U54 AI057159/AI/NIAID NIH HHS/United States
- DP1OD003958-01/OD/NIH HHS/United States
- DP1 CA174427/CA/NCI NIH HHS/United States
- P50 HG006193/HG/NHGRI NIH HHS/United States
- 1P50HG006193-01/HG/NHGRI NIH HHS/United States
- DP1 OD003958/OD/NIH HHS/United States
- DP2 OD002230/OD/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- DP1 DA035083/DA/NIDA NIH HHS/United States
- DP1 OD003893/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

