Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation
- PMID: 23236167
- PMCID: PMC3535655
- DOI: 10.1073/pnas.1210371110
Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation
Abstract
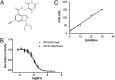
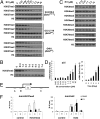
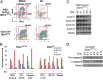
Ezh2 (Enhancer of zeste homolog 2) protein is the enzymatic component of the Polycomb repressive complex 2 (PRC2), which represses gene expression by methylating lysine 27 of histone H3 (H3K27) and regulates cell proliferation and differentiation during embryonic development. Recently, hot-spot mutations of Ezh2 were identified in diffused large B-cell lymphomas and follicular lymphomas. To investigate if tumor growth is dependent on the enzymatic activity of Ezh2, we developed a potent and selective small molecule inhibitor, EI1, which inhibits the enzymatic activity of Ezh2 through direct binding to the enzyme and competing with the methyl group donor S-Adenosyl methionine. EI1-treated cells exhibit genome-wide loss of H3K27 methylation and activation of PRC2 target genes. Furthermore, inhibition of Ezh2 by EI1 in diffused large B-cell lymphomas cells carrying the Y641 mutations results in decreased proliferation, cell cycle arrest, and apoptosis. These results provide strong validation of Ezh2 as a potential therapeutic target for the treatment of cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. - PubMed
-
- Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. - PubMed
-
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35(6):323–332. - PubMed
-
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–570. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

