The evolution of lineage-specific regulatory activities in the human embryonic limb
- PMID: 23827682
- PMCID: PMC3785101
- DOI: 10.1016/j.cell.2013.05.056
The evolution of lineage-specific regulatory activities in the human embryonic limb
Abstract
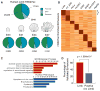
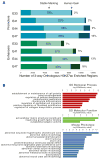
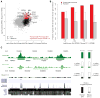
The evolution of human anatomical features likely involved changes in gene regulation during development. However, the nature and extent of human-specific developmental regulatory functions remain unknown. We obtained a genome-wide view of cis-regulatory evolution in human embryonic tissues by comparing the histone modification H3K27ac, which provides a quantitative readout of promoter and enhancer activity, during human, rhesus, and mouse limb development. Based on increased H3K27ac, we find that 13% of promoters and 11% of enhancers have gained activity on the human lineage since the human-rhesus divergence. These gains largely arose by modification of ancestral regulatory activities in the limb or potential co-option from other tissues and are likely to have heterogeneous genetic causes. Most enhancers that exhibit gain of activity in humans originated in mammals. Gains at promoters and enhancers in the human limb are associated with increased gene expression, suggesting they include molecular drivers of human morphological evolution.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Evolutionary biology: The handiwork of tinkering.Nature. 2013 Aug 8;500(7461):158-9. doi: 10.1038/500158a. Nature. 2013. PMID: 23925235 No abstract available.
References
-
- Aiello L, Dean C. An Introduction to Human Evolutionary Anatomy. San Diego: Academic Press; 2002.
-
- Bramble DM, Lieberman DE. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. - PubMed
-
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

