Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche
- PMID: 23467088
- PMCID: PMC3982379
- DOI: 10.1038/nature11979
Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche
Abstract
Epithelial ovarian cancer (EOC) is the fifth leading cause of cancer deaths among women in the United States, but its pathogenesis is poorly understood. Some epithelial cancers are known to occur in transitional zones between two types of epithelium, whereas others have been shown to originate in epithelial tissue stem cells. The stem cell niche of the ovarian surface epithelium (OSE), which is ruptured and regenerates during ovulation, has not yet been defined unequivocally. Here we identify the hilum region of the mouse ovary, the transitional (or junction) area between the OSE, mesothelium and tubal (oviductal) epithelium, as a previously unrecognized stem cell niche of the OSE. We find that cells of the hilum OSE are cycling slowly and express stem and/or progenitor cell markers ALDH1, LGR5, LEF1, CD133 and CK6B. These cells display long-term stem cell properties ex vivo and in vivo, as shown by our serial sphere generation and long-term lineage-tracing assays. Importantly, the hilum cells show increased transformation potential after inactivation of tumour suppressor genes Trp53 and Rb1, whose pathways are altered frequently in the most aggressive and common type of human EOC, high-grade serous adenocarcinoma. Our study supports experimentally the idea that susceptibility of transitional zones to malignant transformation may be explained by the presence of stem cell niches in those areas. Identification of a stem cell niche for the OSE may have important implications for understanding EOC pathogenesis.
Figures
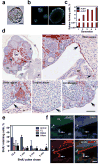
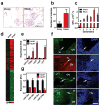

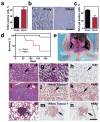
Comment in
-
Stem cells: Anatomy of an ovarian cancer.Nature. 2013 Mar 14;495(7440):183-4. doi: 10.1038/nature11962. Epub 2013 Mar 6. Nature. 2013. PMID: 23467086 No abstract available.
-
Ovarian cancer: At the starting line.Nat Rev Cancer. 2013 May;13(5):296-7. doi: 10.1038/nrc3510. Epub 2013 Mar 28. Nat Rev Cancer. 2013. PMID: 23535843 No abstract available.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
-
- Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–808. - PubMed
-
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. - PubMed
-
- Auersperg N. The origin of ovarian carcinomas: a unifying hypothesis. Int J Gynecol Pathol. 2011;30:12–21. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

