Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization
- PMID: 24591624
- PMCID: PMC3986200
- DOI: 10.1073/pnas.1400975111
Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization
Abstract
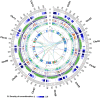
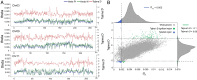
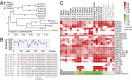
As an economic crop, pepper satisfies people's spicy taste and has medicinal uses worldwide. To gain a better understanding of Capsicum evolution, domestication, and specialization, we present here the genome sequence of the cultivated pepper Zunla-1 (C. annuum L.) and its wild progenitor Chiltepin (C. annuum var. glabriusculum). We estimate that the pepper genome expanded ∼0.3 Mya (with respect to the genome of other Solanaceae) by a rapid amplification of retrotransposons elements, resulting in a genome comprised of ∼81% repetitive sequences. Approximately 79% of 3.48-Gb scaffolds containing 34,476 protein-coding genes were anchored to chromosomes by a high-density genetic map. Comparison of cultivated and wild pepper genomes with 20 resequencing accessions revealed molecular footprints of artificial selection, providing us with a list of candidate domestication genes. We also found that dosage compensation effect of tandem duplication genes probably contributed to the pungent diversification in pepper. The Capsicum reference genome provides crucial information for the study of not only the evolution of the pepper genome but also, the Solanaceae family, and it will facilitate the establishment of more effective pepper breeding programs.
Keywords: Solanaceae evolution; de novo genome sequence; genome expansion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Evolution of a hot genome.Proc Natl Acad Sci U S A. 2014 Apr 8;111(14):5069-70. doi: 10.1073/pnas.1402378111. Epub 2014 Mar 27. Proc Natl Acad Sci U S A. 2014. PMID: 24706896 Free PMC article. No abstract available.
References
-
- Moscone EA, et al. The evolution of chili peppers (Capsicum – Solanaceae): A cytogenetic perspective. Acta Hortic. 2007;745:137–170.
-
- Perry L, et al. Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the Americas. Science. 2007;315(5814):986–988. - PubMed
-
- Bosland PW, Votava EJ. Peppers: Vegetable and spice Capsicums. Crops Prod Sci Hortic. 2012;12:1–11.
-
- Hayman M, Kam PCA. Capsaicin: A review of its pharmacology and clinical applications. Curr Anaesth Crit Care. 2008;19(5-6):338–343.
-
- Votava E, Nabhan G, Bosland P. Genetic diversity and similarity revealed via molecular analysis among and within an in situ population and ex situ accessions of chiltepín (Capsicum annuum var. glabriusculum) Conserv Genet. 2002;3(2):123–129.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

