Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A
- PMID: 23658530
- PMCID: PMC3642083
- DOI: 10.1371/journal.pgen.1003489
Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A
Abstract
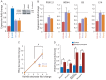
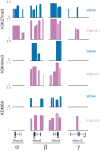
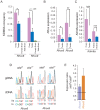
The Rhox cluster on the mouse X chromosome contains reproduction-related homeobox genes expressed in a sexually dimorphic manner. We report that two members of the Rhox cluster, Rhox6 and 9, are regulated by de-methylation of histone H3 at lysine 27 by KDM6A, a histone demethylase with female-biased expression. Consistent with other homeobox genes, Rhox6 and 9 are in bivalent domains prior to embryonic stem cell differentiation and thus poised for activation. In female mouse ES cells, KDM6A is specifically recruited to Rhox6 and 9 for gene activation, a process inhibited by Kdm6a knockdown in a dose-dependent manner. In contrast, KDM6A occupancy at Rhox6 and 9 is low in male ES cells and knockdown has no effect on expression. In mouse ovary where Rhox6 and 9 remain highly expressed, KDM6A occupancy strongly correlates with expression. Our study implicates Kdm6a, a gene that escapes X inactivation, in the regulation of genes important in reproduction, suggesting that KDM6A may play a role in the etiology of developmental and reproduction-related effects of X chromosome anomalies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Wang J, Mager J, Schnedier E, Magnuson T (2002) The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome 13: 493–503. - PubMed
-
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694. - PubMed
-
- Swigut T, Wysocka J (2007) H3K27 demethylases, at long last. Cell 131: 29–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

