Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state
- PMID: 23869022
- PMCID: PMC3959726
- DOI: 10.1126/science.1238303
Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state
Abstract
The ribosome is centrally situated to sense metabolic states, but whether its activity, in turn, coherently rewires transcriptional responses is unknown. Here, through integrated chemical-genetic analyses, we found that a dominant transcriptional effect of blocking protein translation in cancer cells was inactivation of heat shock factor 1 (HSF1), a multifaceted transcriptional regulator of the heat-shock response and many other cellular processes essential for anabolic metabolism, cellular proliferation, and tumorigenesis. These analyses linked translational flux to the regulation of HSF1 transcriptional activity and to the modulation of energy metabolism. Targeting this link with translation initiation inhibitors such as rocaglates deprived cancer cells of their energy and chaperone armamentarium and selectively impaired the proliferation of both malignant and premalignant cells with early-stage oncogenic lesions.
Figures



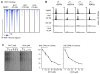
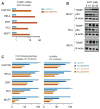
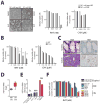
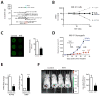
Comment in
-
Cell biology. Trans-HSF1 express.Science. 2013 Jul 19;341(6143):242-3. doi: 10.1126/science.1242359. Science. 2013. PMID: 23869008 No abstract available.
-
HSF1 in Translation.Cancer Cell. 2013 Aug 12;24(2):147-9. doi: 10.1016/j.ccr.2013.07.017. Cancer Cell. 2013. PMID: 23948296
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

