A snapshot of the hepatic transcriptome: ad libitum alcohol intake suppresses expression of cholesterol synthesis genes in alcohol-preferring (P) rats
- PMID: 25542004
- PMCID: PMC4277277
- DOI: 10.1371/journal.pone.0110501
A snapshot of the hepatic transcriptome: ad libitum alcohol intake suppresses expression of cholesterol synthesis genes in alcohol-preferring (P) rats
Abstract
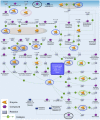
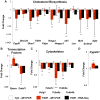
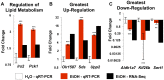
Research is uncovering the genetic and biochemical effects of consuming large quantities of alcohol. One prime example is the J- or U-shaped relationship between the levels of alcohol consumption and the risk of atherosclerotic cardiovascular disease. Moderate alcohol consumption in humans (about 30 g ethanol/d) is associated with reduced risk of coronary heart disease, while abstinence and heavier alcohol intake is linked to increased risk. However, the hepatic consequences of moderate alcohol drinking are largely unknown. Previous data from alcohol-preferring (P) rats showed that chronic consumption does not produce significant hepatic steatosis in this well-established model. Therefore, free-choice alcohol drinking in P rats may mimic low risk or nonhazardous drinking in humans, and chronic exposure in P animals can illuminate the molecular underpinnings of free-choice drinking in the liver. To address this gap, we captured the global, steady-state liver transcriptome following a 23 week free-choice, moderate alcohol consumption regimen (∼ 7.43 g ethanol/kg/day) in inbred alcohol-preferring (iP10a) rats. Chronic consumption led to down-regulation of nine genes in the cholesterol biosynthesis pathway, including HMG-CoA reductase, the rate-limiting step for cholesterol synthesis. These findings corroborate our phenotypic analyses, which indicate that this paradigm produced animals whose hepatic triglyceride levels, cholesterol levels and liver histology were indistinguishable from controls. These findings explain, at least in part, the J- or U-shaped relationship between cardiovascular risk and alcohol intake, and provide outstanding candidates for future studies aimed at understanding the mechanisms that underlie the salutary cardiovascular benefits of chronic low risk and nonhazardous alcohol intake.
Conflict of interest statement
Figures

References
-
- Lieber CS, DeCarli LM (1989) Liquid diet technique of ethanol administration: 1989 update. Alcohol Alcohol 24:197–211. - PubMed
-
- Tsukamoto H, Reidelberger RD, French SW, Largman C (1984) Long-term cannulation model for blood sampling and intragastric infusion in the rat. Am J Physiol 247:R595–599. - PubMed
-
- de la M Hall P, Lieber CS, DeCarli LM, French SW, Lindros KO, et al. (2001) Models of alcoholic liver disease in rodents: a critical evaluation. Alcohol Clin Exp Res 25:254S–261S. - PubMed
-
- Kannel WB, Ellison RC (1996) Alcohol and coronary heart disease: the evidence for a protective effect. Clin Chim Acta 246:59–76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

