Induction of a hemogenic program in mouse fibroblasts
- PMID: 23770078
- PMCID: PMC3735774
- DOI: 10.1016/j.stem.2013.05.024
Induction of a hemogenic program in mouse fibroblasts
Abstract
Definitive hematopoiesis emerges during embryogenesis via an endothelial-to-hematopoietic transition. We attempted to induce this process in mouse fibroblasts by screening a panel of factors for hemogenic activity. We identified a combination of four transcription factors, Gata2, Gfi1b, cFos, and Etv6, that efficiently induces endothelial-like precursor cells, with the subsequent appearance of hematopoietic cells. The precursor cells express a human CD34 reporter, Sca1, and Prominin1 within a global endothelial transcription program. Emergent hematopoietic cells possess nascent hematopoietic stem cell gene-expression profiles and cell-surface phenotypes. After transgene silencing and reaggregation culture, the specified cells generate hematopoietic colonies in vitro. Thus, we show that a simple combination of transcription factors is sufficient to induce a complex, dynamic, and multistep developmental program in vitro. These findings provide insights into the specification of definitive hemogenesis and a platform for future development of patient-specific stem and progenitor cells, as well as more-differentiated blood products.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
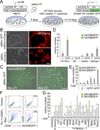
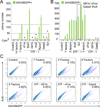

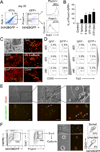



Comment in
-
From skin to blood: a new member joins the iClub.Cell Stem Cell. 2013 Aug 1;13(2):131-3. doi: 10.1016/j.stem.2013.07.007. Cell Stem Cell. 2013. PMID: 23910075
-
"There will be blood" from fibroblasts.Cell Cycle. 2014;13(3):335-6. doi: 10.4161/cc.27507. Epub 2013 Dec 13. Cell Cycle. 2014. PMID: 24335410 Free PMC article. No abstract available.
References
-
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. - PubMed
-
- Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24:1249–1257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

