De novo copy number variations in cloned dogs from the same nuclear donor
- PMID: 24313905
- PMCID: PMC3878922
- DOI: 10.1186/1471-2164-14-863
De novo copy number variations in cloned dogs from the same nuclear donor
Abstract
Background: Somatic mosaicism of copy number variants (CNVs) in human body organs and de novo CNV event in monozygotic twins suggest that de novo CNVs can occur during mitotic recombination. These de novo CNV events are important for understanding genetic background of evolution and diverse phenotypes. In this study, we explored de novo CNV event in cloned dogs with identical genetic background.
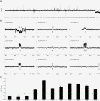
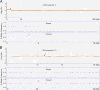
Results: We analyzed CNVs in seven cloned dogs using the nuclear donor genome as reference by array-CGH, and identified five de novo CNVs in two of the seven clones. Genomic qPCR, dye-swap array-CGH analysis and B-allele profile analysis were used for their validation. Two larger de novo CNVs (5.2 Mb and 338 Kb) on chromosomes X and 19 in clone-3 were consistently validated by all three experiments. The other three smaller CNVs (sized from 36.1 to 76.4 Kb) on chromosomes 2, 15 and 32 in clone-3 and clone-6 were verified by at least one of the three validations. In addition to the de novo CNVs, we identified a 37 Mb-sized copy neutral de novo loss of heterozygosity event on chromosome 2 in clone-6.
Conclusions: To our knowledge, this is the first report of de novo CNVs in the cloned dogs which were generated by somatic cell nuclear transfer technology. To study de novo genetic events in cloned animals can help understand formation mechanisms of genetic variants and their biological implications.
Figures



References
-
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S. et al.Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;14:803–819. doi: 10.1038/nature04338. - DOI - PubMed
-
- Piotrowski A, Bruder CE, Andersson R, Diaz de Ståhl T, Menzel U, Sandgren J, Poplawski A, von Tell D, Crasto C, Bogdan A, Bartoszewski R, Bebok Z, Krzyzanowski M, Jankowski Z, Partridge EC, Komorowski J, Dumanski JP. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;14:1118–1124. doi: 10.1002/humu.20815. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

