Cycles in spatial and temporal chromosomal organization driven by the circadian clock
- PMID: 24056944
- PMCID: PMC3885543
- DOI: 10.1038/nsmb.2667
Cycles in spatial and temporal chromosomal organization driven by the circadian clock
Abstract
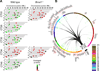
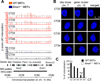
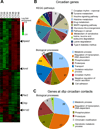
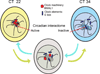
Dynamic transitions in the epigenome have been associated with regulated patterns of nuclear organization. The accumulating evidence that chromatin remodeling is implicated in circadian function prompted us to explore whether the clock may control nuclear architecture. We applied the chromosome conformation capture on chip technology in mouse embryonic fibroblasts (MEFs) to demonstrate the presence of circadian long-range interactions using the clock-controlled Dbp gene as bait. The circadian genomic interactions with Dbp were highly specific and were absent in MEFs whose clock was disrupted by ablation of the Bmal1 gene (also called Arntl). We establish that the Dbp circadian interactome contains a wide variety of genes and clock-related DNA elements. These findings reveal a previously unappreciated circadian and clock-dependent shaping of the nuclear landscape.
Figures


References
-
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. - PubMed
-
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. - PubMed
-
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annual review of genetics. 2006;40:409–448. - PubMed
-
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. - PubMed
METHODS-ONLY REFERENCES
-
- Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. - PubMed
-
- Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–117. - PubMed
-
- Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

