14-3-3ζ turns TGF-β's function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2
- PMID: 25670079
- PMCID: PMC4325275
- DOI: 10.1016/j.ccell.2014.11.025
14-3-3ζ turns TGF-β's function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2
Abstract
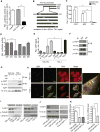
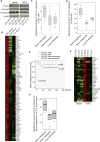
Transforming growth factor β (TGF-β) functions as a tumor suppressor in premalignant cells but as a metastasis promoter in cancer cells. The dichotomous functions of TGF-β are proposed to be dictated by different partners of its downstream effector Smads. However, the mechanism for the contextual changes of Smad partners remained undefined. Here, we demonstrate that 14-3-3ζ destabilizes p53, a Smad partner in premalignant mammary epithelial cells, by downregulating 14-3-3σ, thus turning off TGF-β's tumor suppression function. Conversely, 14-3-3ζ stabilizes Gli2 in breast cancer cells, and Gli2 partners with Smads to activate PTHrP and promote TGF-β-induced bone metastasis. The 14-3-3ζ-driven contextual changes of Smad partners from p53 to Gli2 may serve as biomarkers and therapeutic targets of TGF-β-mediated cancer progression.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Comment in
-
14-3-3ζ turns TGF-β to the dark side.Cancer Cell. 2015 Feb 9;27(2):151-3. doi: 10.1016/j.ccell.2015.01.005. Cancer Cell. 2015. PMID: 25670073
-
Tipping the balance between good and evil: aberrant 14-3-3ζ expression drives oncogenic TGF-β signaling in metastatic breast cancers.Breast Cancer Res. 2015 Jul 11;17:92. doi: 10.1186/s13058-015-0603-2. Breast Cancer Res. 2015. PMID: 26160166 Free PMC article.
References
-
- Akahira J, Sugihashi Y, Suzuki T, Ito K, Niikura H, Moriya T, Nitta M, Okamura H, Inoue S, Sasano H, et al. Decreased expression of 14-3-3 sigma is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin Cancer Res. 2004;10:2687–2693. - PubMed
-
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

