Reexpression of LSAMP inhibits tumor growth in a preclinical osteosarcoma model
- PMID: 24885297
- PMCID: PMC4029956
- DOI: 10.1186/1476-4598-13-93
Reexpression of LSAMP inhibits tumor growth in a preclinical osteosarcoma model
Abstract
Background: Osteosarcomas are the most common primary malignant tumors of bone, showing complex chromosomal rearrangements with multiple gains and losses. A frequent deletion within the chromosomal region 3q13.31 has been identified by us and others, and is mainly reported to be present in osteosarcomas. The purpose of the study was to further characterize the frequency and the extent of the deletion in an extended panel of osteosarcoma samples, and the expression level of the affected genes within the region. We have identified LSAMP as the target gene for the deletion, and have studied the functional implications of LSAMP-reexpression.
Methods: LSAMP copy number, expression level and protein level were investigated by quantitative PCR and western blotting in an osteosarcoma panel. The expression of LSAMP was restored in an osteosarcoma cell line, and differences in proliferation rate, tumor formation, gene expression, migration rate, differentiation capabilities, cell cycle distribution and apoptosis were investigated by metabolic dyes, tumor formation in vivo, gene expression profiling, time-lapse photography, differentiation techniques and flow cytometry, respectively.
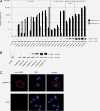
Results: We found reduced copy number of LSAMP in 45/76 osteosarcoma samples, reduced expression level in 25/42 samples and protein expression in 9/42 samples. By restoring the expression of LSAMP in a cell line with a homozygous deletion of the gene, the proliferation rate in vitro was significantly reduced and tumor growth in vivo was significantly delayed. In response to reexpression of LSAMP, mRNA expression profiling revealed consistent upregulation of the genes hairy and enhancer of split 1 (HES1), cancer/testis antigen 2 (CTAG2) and kruppel-like factor 10 (KLF10).
Conclusions: The high frequency and the specificity of the deletion indicate that it is important for the development of osteosarcomas. The deletion targets the tumor suppressor LSAMP, and based on the functional evidence, the tumor suppressor function of LSAMP is most likely exerted by reducing the proliferation rate of the tumor cells, possibly by indirectly upregulating one or more of the genes HES1, CTAG2 or KLF10. To our knowledge, this study describes novel functions of LSAMP, a first step to understanding the functional role of this specific deletion in osteosarcomas.
Figures


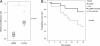

References
-
- Fletcher CDM, Unni KK, Mertens F. World Health Organization Classification of Tumours. Pathology and Genetics: Tumours of Soft Tissue and Bone. Lyon: IARCPress; 2002.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

