Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis
- PMID: 17000729
- PMCID: PMC1698089
- DOI: 10.1128/IAI.01250-06
Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis
Abstract
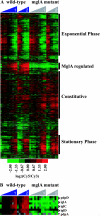
The facultative intracellular bacterium Francisella tularensis causes the zoonotic disease tularemia. F. tularensis resides within host macrophages in vivo, and this ability is essential for pathogenesis. The transcription factor MglA is required for the expression of several Francisella genes that are necessary for replication in macrophages and for virulence in mice. We hypothesized that the identification of MglA-regulated genes in the Francisella genome by transcriptional profiling of wild-type and mglA mutant bacteria would lead to the discovery of new virulence factors utilized by F. tularensis. A total of 102 MglA-regulated genes were identified, the majority of which were positively regulated, including all of the Francisella pathogenicity island (FPI) genes. We mutated novel MglA-regulated genes and tested the mutants for their ability to replicate and induce cytotoxicity in macrophages and to grow in mice. Mutations in MglA-regulated genes within the FPI (pdpB and cds2) as well as outside the FPI (FTT0989, oppB, and FTT1209c) were either attenuated or hypervirulent in macrophages compared to the wild-type strain. All of these mutants exhibited decreased fitness in vivo in competition experiments with wild-type bacteria. We have identified five new Francisella virulence genes, and our results suggest that characterizations of additional MglA-regulated genes will yield further insights into the pathogenesis of this bacterium.
Figures


References
-
- Ansai, T., W. Yu, S. Urnowey, S. Barik, and T. Takehara. 2003. Construction of a pepO gene-deficient mutant of Porphyromonas gingivalis: potential role of endopeptidase O in the invasion of host cells. Oral Microbiol. Immunol. 18:398-400. - PubMed
-
- Anthony, L. S., M. Z. Gu, S. C. Cowley, W. W. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 137:2697-2703. - PubMed
-
- Awano, S., T. Ansai, H. Mochizuki, W. Yu, K. Tanzawa, A. J. Turner, and T. Takehara. 1999. Sequencing, expression and biochemical characterization of the Porphyromonas gingivalis pepO gene encoding a protein homologous to human endothelin-converting enzyme. FEBS Lett. 460:139-144. - PubMed
-
- Baron, G. S., and F. E. Nano. 1999. An erythromycin resistance cassette and mini-transposon for constructing transcriptional fusions to cat. Gene 229:59-65. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

