Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population
- PMID: 25850675
- PMCID: PMC4390738
- DOI: 10.1016/j.devcel.2015.01.025
Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population
Abstract
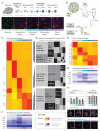
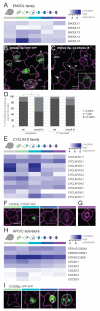
Developmental transitions can be described in terms of morphology and the roles of individual genes, but also in terms of global transcriptional and epigenetic changes. Temporal dissections of transcriptome changes, however, are rare for intact, developing tissues. We used RNA sequencing and microarray platforms to quantify gene expression from labeled cells isolated by fluorescence-activated cell sorting to generate cell-type-specific transcriptomes during development of an adult stem-cell lineage in the Arabidopsis leaf. We show that regulatory modules in this early lineage link cell types that had previously been considered to be under separate control and provide evidence for recruitment of individual members of gene families for different developmental decisions. Because stomata are physiologically important and because stomatal lineage cells exhibit exemplary division, cell fate, and cell signaling behaviors, this dataset serves as a valuable resource for further investigations of fundamental developmental processes.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


References
-
- Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. - PubMed
-
- Birnbaum K, Shasha DE, Wang JYY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

