Understanding multicellular function and disease with human tissue-specific networks
- PMID: 25915600
- PMCID: PMC4828725
- DOI: 10.1038/ng.3259
Understanding multicellular function and disease with human tissue-specific networks
Abstract
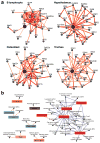
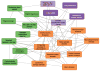
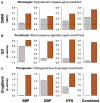
Tissue and cell-type identity lie at the core of human physiology and disease. Understanding the genetic underpinnings of complex tissues and individual cell lineages is crucial for developing improved diagnostics and therapeutics. We present genome-wide functional interaction networks for 144 human tissues and cell types developed using a data-driven Bayesian methodology that integrates thousands of diverse experiments spanning tissue and disease states. Tissue-specific networks predict lineage-specific responses to perturbation, identify the changing functional roles of genes across tissues and illuminate relationships among diseases. We introduce NetWAS, which combines genes with nominally significant genome-wide association study (GWAS) P values and tissue-specific networks to identify disease-gene associations more accurately than GWAS alone. Our webserver, GIANT, provides an interface to human tissue networks through multi-gene queries, network visualization, analysis tools including NetWAS and downloadable networks. GIANT enables systematic exploration of the landscape of interacting genes that shape specialized cellular functions across more than a hundred human tissues and cell types.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
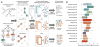

Comment in
-
Molecular networks in context.Nat Biotechnol. 2015 Jul;33(7):720-1. doi: 10.1038/nbt.3283. Nat Biotechnol. 2015. PMID: 26154012 No abstract available.
References
-
- D’Agati VD. The spectrum of focal segmental glomerulosclerosis: new insights. Curr Opin Nephrol Hypertens. 2008;17:271–81. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

