Overlapping DNA methylation dynamics in mouse intestinal cell differentiation and early stages of malignant progression
- PMID: 25933092
- PMCID: PMC4416816
- DOI: 10.1371/journal.pone.0123263
Overlapping DNA methylation dynamics in mouse intestinal cell differentiation and early stages of malignant progression
Abstract
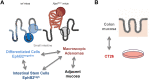
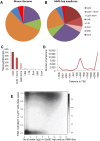
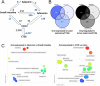
Mouse models of intestinal crypt cell differentiation and tumorigenesis have been used to characterize the molecular mechanisms underlying both processes. DNA methylation is a key epigenetic mark and plays an important role in cell identity and differentiation programs and cancer. To get insights into the dynamics of cell differentiation and malignant transformation we have compared the DNA methylation profiles along the mouse small intestine crypt and early stages of tumorigenesis. Genome-scale analysis of DNA methylation together with microarray gene expression have been applied to compare intestinal crypt stem cells (EphB2high), differentiated cells (EphB2negative), ApcMin/+ adenomas and the corresponding non-tumor adjacent tissue, together with small and large intestine samples and the colon cancer cell line CT26. Compared with late stages, small intestine crypt differentiation and early stages of tumorigenesis display few and relatively small changes in DNA methylation. Hypermethylated loci are largely shared by the two processes and affect the proximities of promoter and enhancer regions, with enrichment in genes associated with the intestinal stem cell signature and the PRC2 complex. The hypermethylation is progressive, with minute levels in differentiated cells, as compared with intestinal stem cells, and reaching full methylation in advanced stages. Hypomethylation shows different signatures in differentiation and cancer and is already present in the non-tumor tissue adjacent to the adenomas in ApcMin/+ mice, but at lower levels than advanced cancers. This study provides a reference framework to decipher the mechanisms driving mouse intestinal tumorigenesis and also the human counterpart.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

