Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue
- PMID: 25436972
- PMCID: PMC4343194
- DOI: 10.1038/ni.3047
Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue
Abstract
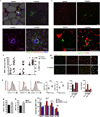
Invariant natural killer T cells (iNKT cells) are lipid-sensing innate T cells that are restricted by the antigen-presenting molecule CD1d and express the transcription factor PLZF. iNKT cells accumulate in adipose tissue, where they are anti-inflammatory, but the factors that contribute to their anti-inflammatory nature, as well as their targets in adipose tissue, are unknown. Here we found that iNKT cells in adipose tissue had a unique transcriptional program and produced interleukin 2 (IL-2) and IL-10. Unlike other iNKT cells, they lacked PLZF but expressed the transcription factor E4BP4, which controlled their IL-10 production. The adipose iNKT cells were a tissue-resident population that induced an anti-inflammatory phenotype in macrophages and, through the production of IL-2, controlled the number, proliferation and suppressor function of regulatory T cells (Treg cells) in adipose tissue. Thus, iNKT cells in adipose tissue are unique regulators of immunological homeostasis in this tissue.
Figures


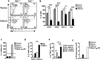



References
-
- Cohen NR, Garg S, Brenner MB. Antigen Presentation by CD1 Lipids, T Cells, and NKT Cells in Microbial Immunity. Adv Immunol. 2009;102:1–94. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- DK057521/DK/NIDDK NIH HHS/United States
- R01 AI113046/AI/NIAID NIH HHS/United States
- AI059739/AI/NIAID NIH HHS/United States
- R01 AI059739/AI/NIAID NIH HHS/United States
- R37 AI028973/AI/NIAID NIH HHS/United States
- R01 AI063428/AI/NIAID NIH HHS/United States
- AI063428/AI/NIAID NIH HHS/United States
- P01 AI112521/AI/NIAID NIH HHS/United States
- AI028973/AI/NIAID NIH HHS/United States
- G1001750/MRC_/Medical Research Council/United Kingdom
- R01 AI083988/AI/NIAID NIH HHS/United States
- P30 CA072720/CA/NCI NIH HHS/United States
- T32 AI049823/AI/NIAID NIH HHS/United States
- R56 AI063428/AI/NIAID NIH HHS/United States
- K08 AI102945/AI/NIAID NIH HHS/United States
- R01 AI028973/AI/NIAID NIH HHS/United States
- P30 DK057521/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

