High-affinity transporters for NAD+ precursors in Candida glabrata are regulated by Hst1 and induced in response to niacin limitation
- PMID: 19451228
- PMCID: PMC2715804
- DOI: 10.1128/MCB.01461-08
High-affinity transporters for NAD+ precursors in Candida glabrata are regulated by Hst1 and induced in response to niacin limitation
Abstract
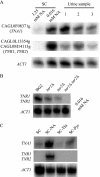
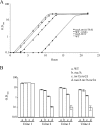
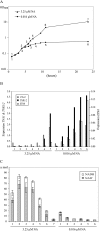
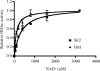
The yeast Candida glabrata is an opportunistic pathogen of humans. C. glabrata is a NAD(+) auxotroph, and its growth depends on the availability of niacin (environmental vitamin precursors of NAD(+)). We have previously shown that a virulence-associated adhesin, encoded by EPA6, is transcriptionally induced in response to niacin limitation. Here we used transcript profiling to characterize the transcriptional response to niacin limitation and the roles of the sirtuins Hst1, Hst2, and Sir2 in mediating this response. The majority of genes transcriptionally induced by niacin limitation are regulated by Hst1, suggesting that it is the primary sensor of niacin limitation in C. glabrata. We show that three highly induced genes, TNA1, TNR1, and TNR2, encode transporters which are necessary and sufficient for high-affinity uptake of NAD(+) precursors. Strikingly, if a tna1 tnr1 tnr2 mutant is starved for niacin, it exhibits an extended lag phase, suggesting a central role for the transporters in restoring NAD(+) homeostasis after niacin limitation. Lastly, we had previously shown that the adhesin encoded by EPA6 is induced during experimental urinary tract infection (UTI); we show here that EPA6 transcriptional induction during UTI is strongly enhanced in the tna1 tnr1 tnr2 mutant strain, implicating the transporters in the growth of C. glabrata during infection.
Figures



References
-
- Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen, S. S. Lin, J. K. Manchester, J. I. Gordon, and D. A. Sinclair. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 27718881-18890. - PubMed
-
- Belenky, P., K. L. Bogan, and C. Brenner. 2007. NAD+ metabolism in health and disease. Trends Biochem. Sci. 3212-19. - PubMed
-
- Belenky, P. A., T. G. Moga, and C. Brenner. 2008. Saccharomyces cerevisiae YOR071C encodes the high affinity nicotinamide riboside transporter Nrt1. J. Biol. Chem. 2838075-8079. - PubMed
-
- Bieganowski, P., and C. Brenner. 2004. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117495-502. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
