Transcriptomic Changes Due to Cytoplasmic TDP-43 Expression Reveal Dysregulation of Histone Transcripts and Nuclear Chromatin
- PMID: 26510133
- PMCID: PMC4624943
- DOI: 10.1371/journal.pone.0141836
Transcriptomic Changes Due to Cytoplasmic TDP-43 Expression Reveal Dysregulation of Histone Transcripts and Nuclear Chromatin
Abstract
TAR DNA-binding protein 43 (TDP-43) is normally a nuclear RNA-binding protein that exhibits a range of functions including regulation of alternative splicing, RNA trafficking, and RNA stability. However, in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP), TDP-43 is abnormally phosphorylated, ubiquitinated, and cleaved, and is mislocalized to the cytoplasm where it forms distinctive aggregates. We previously developed a mouse model expressing human TDP-43 with a mutation in its nuclear localization signal (ΔNLS-hTDP-43) so that the protein preferentially localizes to the cytoplasm. These mice did not exhibit a significant number of cytoplasmic aggregates, but did display dramatic changes in gene expression as measured by microarray, suggesting that cytoplasmic TDP-43 may be associated with a toxic gain-of-function. Here, we analyze new RNA-sequencing data from the ΔNLS-hTDP-43 mouse model, together with published RNA-sequencing data obtained previously from TDP-43 antisense oligonucleotide (ASO) knockdown mice to investigate further the dysregulation of gene expression in the ΔNLS model. This analysis reveals that the transcriptomic effects of the overexpression of the ΔNLS-hTDP-43 transgene are likely due to a gain of cytoplasmic function. Moreover, cytoplasmic TDP-43 expression alters transcripts that regulate chromatin assembly, the nucleolus, lysosomal function, and histone 3' untranslated region (UTR) processing. These transcriptomic alterations correlate with observed histologic abnormalities in heterochromatin structure and nuclear size in transgenic mouse and human brains.
Conflict of interest statement
Figures



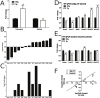
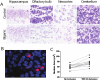
References
-
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001. September 28;276(39):36337–43. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

