SARS-CoV-2 human challenge reveals biomarkers that discriminate early and late phases of respiratory viral infections
- PMID: 39616162
- PMCID: PMC11608262
- DOI: 10.1038/s41467-024-54764-3
SARS-CoV-2 human challenge reveals biomarkers that discriminate early and late phases of respiratory viral infections
Abstract
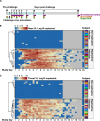
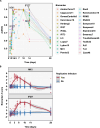
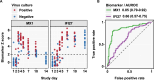
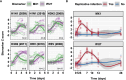
Blood transcriptional biomarkers of acute viral infections typically reflect type 1 interferon (IFN) signalling, but it is not known whether there are biological differences in their regulation that can be leveraged for distinct translational applications. We use high frequency sampling in the SARS-CoV-2 human challenge model to show induction of IFN-stimulated gene (ISG) expression with different temporal and cellular profiles. MX1 gene expression correlates with a rapid and transient wave of ISG expression across all cell types, which may precede PCR detection of replicative infection. Another ISG, IFI27, shows a delayed but sustained response restricted to myeloid cells, attributable to gene and cell-specific epigenetic regulation. These findings are reproducible in experimental and naturally acquired infections with influenza, respiratory syncytial virus and rhinovirus. Blood MX1 expression is superior to IFI27 expression for diagnosis of early infection, as a correlate of viral load and for discrimination of virus culture positivity. Therefore, MX1 expression offers potential to stratify patients for antiviral therapy or infection control interventions. Blood IFI27 expression is superior to MX1 expression for diagnostic accuracy across the time course of symptomatic infection and thereby, offers higher diagnostic yield for respiratory virus infections that incur a delay between transmission and testing.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The Authors declare the following competing interests: In the past 3 years, S.A.T. has received remuneration for scientific advisory board membership from Sanofi, GlaxoSmithKline, Foresite Labs and Qiagen. S.A.T. is a co-founder and holds equity in Transition Bio and Ensocell. From 8 January 2024, S.A.T. has been a part-time employee of GlaxoSmithKline. A.J.M., A.C., M.K., M.M. and A.B. are full time employees at hVIVO Services Ltd. No other authors report any competing interests.
Figures


References
-
- Killingley, B. et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med28, 1031–1041 (2022). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

