Integrator mediates the biogenesis of enhancer RNAs
- PMID: 26308897
- PMCID: PMC4718573
- DOI: 10.1038/nature14906
Integrator mediates the biogenesis of enhancer RNAs
Abstract
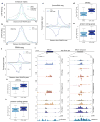
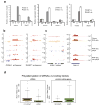
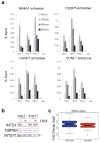
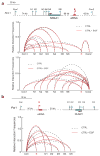
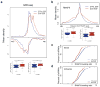
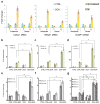
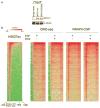
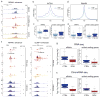
Integrator is a multi-subunit complex stably associated with the carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII). Integrator is endowed with a core catalytic RNA endonuclease activity, which is required for the 3'-end processing of non-polyadenylated, RNAPII-dependent, uridylate-rich, small nuclear RNA genes. Here we examine the requirement of Integrator in the biogenesis of transcripts derived from distal regulatory elements (enhancers) involved in tissue- and temporal-specific regulation of gene expression in metazoans. Integrator is recruited to enhancers and super-enhancers in a stimulus-dependent manner. Functional depletion of Integrator subunits diminishes the signal-dependent induction of enhancer RNAs (eRNAs) and abrogates stimulus-induced enhancer-promoter chromatin looping. Global nuclear run-on and RNAPII profiling reveals a role for Integrator in 3'-end cleavage of eRNA primary transcripts leading to transcriptional termination. In the absence of Integrator, eRNAs remain bound to RNAPII and their primary transcripts accumulate. Notably, the induction of eRNAs and gene expression responsiveness requires the catalytic activity of Integrator complex. We propose a role for Integrator in biogenesis of eRNAs and enhancer function in metazoans.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

