Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity
- PMID: 26406376
- PMCID: PMC4597190
- DOI: 10.1016/j.cell.2015.09.001
Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity
Abstract
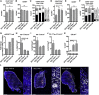
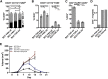
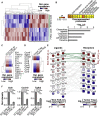
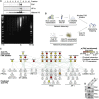
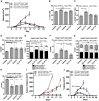
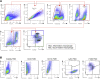
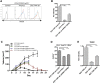
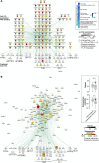
Focal adhesion kinase (FAK) promotes anti-tumor immune evasion. Specifically, the kinase activity of nuclear-targeted FAK in squamous cell carcinoma (SCC) cells drives exhaustion of CD8(+) T cells and recruitment of regulatory T cells (Tregs) in the tumor microenvironment by regulating chemokine/cytokine and ligand-receptor networks, including via transcription of Ccl5, which is crucial. These changes inhibit antigen-primed cytotoxic CD8(+) T cell activity, permitting growth of FAK-expressing tumors. Mechanistically, nuclear FAK is associated with chromatin and exists in complex with transcription factors and their upstream regulators that control Ccl5 expression. Furthermore, FAK's immuno-modulatory nuclear activities may be specific to cancerous squamous epithelial cells, as normal keratinocytes do not have nuclear FAK. Finally, we show that a small-molecule FAK kinase inhibitor, VS-4718, which is currently in clinical development, also drives depletion of Tregs and promotes a CD8(+) T cell-mediated anti-tumor response. Therefore, FAK inhibitors may trigger immune-mediated tumor regression, providing previously unrecognized therapeutic opportunities.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Albasri A., Fadhil W., Scholefield J.H., Durrant L.G., Ilyas M. Nuclear expression of phosphorylated focal adhesion kinase is associated with poor prognosis in human colorectal cancer. Anticancer Res. 2014;34:3969–3974. - PubMed
-
- Beyer M., Schultze J.L. Regulatory T cells in cancer. Blood. 2006;108:804–811. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

