Genome-wide detection of imprinted differentially methylated regions using nanopore sequencing
- PMID: 35787786
- PMCID: PMC9255983
- DOI: 10.7554/eLife.77898
Genome-wide detection of imprinted differentially methylated regions using nanopore sequencing
Abstract
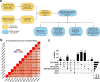
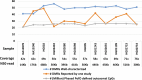
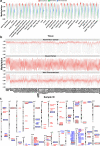
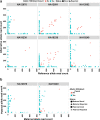
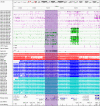
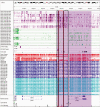
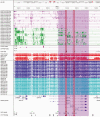
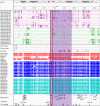
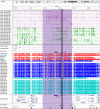
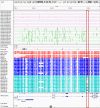
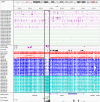
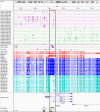
Imprinting is a critical part of normal embryonic development in mammals, controlled by defined parent-of-origin (PofO) differentially methylated regions (DMRs) known as imprinting control regions. Direct nanopore sequencing of DNA provides a means to detect allelic methylation and to overcome the drawbacks of methylation array and short-read technologies. Here, we used publicly available nanopore sequencing data for 12 standard B-lymphocyte cell lines to acquire the genome-wide mapping of imprinted intervals in humans. Using the sequencing data, we were able to phase 95% of the human methylome and detect 94% of the previously well-characterized, imprinted DMRs. In addition, we found 42 novel imprinted DMRs (16 germline and 26 somatic), which were confirmed using whole-genome bisulfite sequencing (WGBS) data. Analysis of WGBS data in mouse (Mus musculus), rhesus monkey (Macaca mulatta), and chimpanzee (Pan troglodytes) suggested that 17 of these imprinted DMRs are conserved. Some of the novel imprinted intervals are within or close to imprinted genes without a known DMR. We also detected subtle parental methylation bias, spanning several kilobases at seven known imprinted clusters. At these blocks, hypermethylation occurs at the gene body of expressed allele(s) with mutually exclusive H3K36me3 and H3K27me3 allelic histone marks. These results expand upon our current knowledge of imprinting and the potential of nanopore sequencing to identify imprinting regions using only parent-offspring trios, as opposed to the large multi-generational pedigrees that have previously been required.
Keywords: DNA methylation; H3K27me3; H3K36me3; allele-specific methylation; computational biology; genetics; genomics; human; imprinting; nanopore sequencing; systems biology.
© 2022, Akbari et al.
Conflict of interest statement
VA, JG, KO, PP, RM, MM, MH, SJ No competing interests declared
Figures



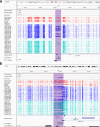
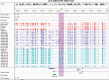
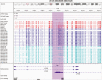
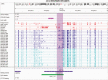
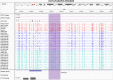


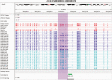
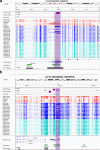



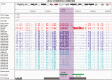






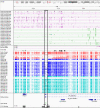
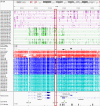
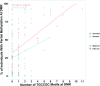
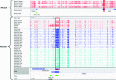

References
-
- Akbari V. NanoMethPhase. swh:1:rev:1657f7aed60604aa7c7f3e77d992d76bee6bf6d3Software Heritage. 2022 https://archive.softwareheritage.org/swh:1:dir:544b123669c23565347328349...
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- SRA/SRP030041
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

