Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons
- PMID: 31746735
- PMCID: PMC6894925
- DOI: 10.7554/eLife.48718
Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons
Abstract
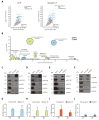
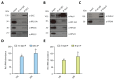
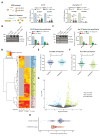
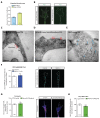
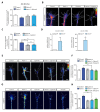
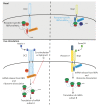
Extrinsic cues trigger the local translation of specific mRNAs in growing axons via cell surface receptors. The coupling of ribosomes to receptors has been proposed as a mechanism linking signals to local translation but it is not known how broadly this mechanism operates, nor whether it can selectively regulate mRNA translation. We report that receptor-ribosome coupling is employed by multiple guidance cue receptors and this interaction is mRNA-dependent. We find that different receptors associate with distinct sets of mRNAs and RNA-binding proteins. Cue stimulation of growing Xenopus retinal ganglion cell axons induces rapid dissociation of ribosomes from receptors and the selective translation of receptor-specific mRNAs. Further, we show that receptor-ribosome dissociation and cue-induced selective translation are inhibited by co-exposure to translation-repressive cues, suggesting a novel mode of signal integration. Our findings reveal receptor-specific interactomes and suggest a generalizable model for cue-selective control of the local proteome.
Keywords: RNA-binding protein; axon; developmental biology; guidance receptor; human; local protein synthesis; mRNA; neuroscience; retinal ganglion cell; xenopus.
© 2019, Koppers et al.
Conflict of interest statement
MK, RC, TS, LW, PV, JQ, SZ, MJ, AD, MM, AB, CK, WH, JF, CH No competing interests declared
Figures




References
-
- Bellon A, Iyer A, Bridi S, Lee FCY, Ovando-Vázquez C, Corradi E, Longhi S, Roccuzzo M, Strohbuecker S, Naik S, Sarkies P, Miska E, Abreu-Goodger C, Holt CE, Baudet ML. miR-182 regulates Slit2-Mediated axon guidance by modulating the local translation of a specific mRNA. Cell Reports. 2017;18:1171–1186. doi: 10.1016/j.celrep.2016.12.093. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

