A computational approach to estimate interorgan metabolic transport in a mammal
- PMID: 24971892
- PMCID: PMC4074118
- DOI: 10.1371/journal.pone.0100963
A computational approach to estimate interorgan metabolic transport in a mammal
Abstract
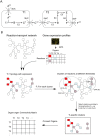
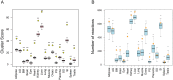
In multicellular organisms metabolism is distributed across different organs, each of which has specific requirements to perform its own specialized task. But different organs also have to support the metabolic homeostasis of the organism as a whole by interorgan metabolite transport. Recent studies have successfully reconstructed global metabolic networks in tissues and cell types and attempts have been made to connect organs with interorgan metabolite transport. Instead of these complicated approaches to reconstruct global metabolic networks, we proposed in this study a novel approach to study interorgan metabolite transport focusing on transport processes mediated by solute carrier (Slc) transporters and their couplings to cognate enzymatic reactions. We developed a computational approach to identify and score potential interorgan metabolite transports based on the integration of metabolism and transports in different organs in the adult mouse from quantitative gene expression data. This allowed us to computationally estimate the connectivity between 17 mouse organs via metabolite transport. Finally, by applying our method to circadian metabolism, we showed that our approach can shed new light on the current understanding of interorgan metabolite transport at a whole-body level in mammals.
Conflict of interest statement
Figures




References
-
- Schilling CH, Letscher D, Palsson BO (2000) Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J Theor Biol 203: 229–248. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

