Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs
- PMID: 22179699
- PMCID: PMC3261569
- DOI: 10.1038/emboj.2011.458
Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs
Abstract
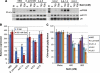
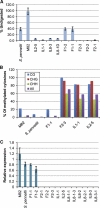
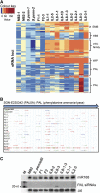
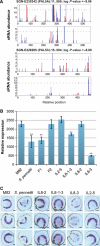
Hybrid organisms may fail to develop, be sterile or they may be more vigorous than either of the parents. Examples of hybrid vigour or hybrid necrosis in the F1 are often not inherited stably in subsequent generations if they are associated with overdominance. There can also be transgressive phenotypes that are inherited stably in these later generations, but the underlying mechanisms are not well understood. Here we have investigated the possibility that stable transgressive phenotypes in the progeny of crosses between cultivated tomato (Solanum lycopersicum cv. M82) and a wild relative (Solanum pennellii, accession LA716) are associated with micro or small interfering(si) RNAs. We identified loci from which these small(s)RNAs were more abundant in hybrids than in either parent and we show that accumulation of such transgressive sRNAs correlated with suppression of the corresponding target genes. In one instance this effect was associated with hypermethylation of the corresponding genomic DNA. Our results illustrate a potential role of transgressive sRNAs in plant breeding and in natural evolution with wild plants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Comment in
-
On epigenetics and epistasis: hybrids and their non-additive interactions.EMBO J. 2012 Jan 18;31(2):249-50. doi: 10.1038/emboj.2011.473. Epub 2011 Dec 23. EMBO J. 2012. PMID: 22193717 Free PMC article.
-
Small RNAs: Little transgressions.Nat Rev Genet. 2012 Jan 10;13(2):75. doi: 10.1038/nrg3155. Nat Rev Genet. 2012. PMID: 22230818 No abstract available.
References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during Trans-Acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Anders S, Wolfgang H (2010) Differential expression analysis for sequence count data. Nat Precedings, http://hdl.handle.net/10101/npre.2010.4282.2 - PMC - PubMed
-
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

