Multiplex, single-cell CRISPRa screening for cell type specific regulatory elements
- PMID: 39294132
- PMCID: PMC11411074
- DOI: 10.1038/s41467-024-52490-4
Multiplex, single-cell CRISPRa screening for cell type specific regulatory elements
Abstract
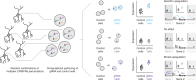
CRISPR-based gene activation (CRISPRa) is a strategy for upregulating gene expression by targeting promoters or enhancers in a tissue/cell-type specific manner. Here, we describe an experimental framework that combines highly multiplexed perturbations with single-cell RNA sequencing (sc-RNA-seq) to identify cell-type-specific, CRISPRa-responsive cis-regulatory elements and the gene(s) they regulate. Random combinations of many gRNAs are introduced to each of many cells, which are then profiled and partitioned into test and control groups to test for effect(s) of CRISPRa perturbations of both enhancers and promoters on the expression of neighboring genes. Applying this method to a library of 493 gRNAs targeting candidate cis-regulatory elements in both K562 cells and iPSC-derived excitatory neurons, we identify gRNAs capable of specifically upregulating intended target genes and no other neighboring genes within 1 Mb, including gRNAs yielding upregulation of six autism spectrum disorder (ASD) and neurodevelopmental disorder (NDD) risk genes in neurons. A consistent pattern is that the responsiveness of individual enhancers to CRISPRa is restricted by cell type, implying a dependency on either chromatin landscape and/or additional trans-acting factors for successful gene activation. The approach outlined here may facilitate large-scale screens for gRNAs that activate genes in a cell type-specific manner.
© 2024. The Author(s).
Conflict of interest statement
S.J.S. receives research funding from BioMarin Pharmaceutical Incorporated. N.A. is the cofounder and on the scientific advisory board of Regel Therapeutics and receives funding from BioMarin Pharmaceutical Incorporated. J.S. is a scientific advisory board member, consultant and/or co-founder of Cajal Neuroscience, Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Phase Genomics, Adaptive Biotechnologies, Scale Biosciences, Sixth Street Capital, Prime Medicine, Somite Therapeutics and Pacific Biosciences. All other authors declare no competing interests.
Figures


Update of
-
Multiplex, single-cell CRISPRa screening for cell type specific regulatory elements.bioRxiv [Preprint]. 2024 Apr 30:2023.03.28.534017. doi: 10.1101/2023.03.28.534017. bioRxiv. 2024. Update in: Nat Commun. 2024 Sep 18;15(1):8209. doi: 10.1038/s41467-024-52490-4. PMID: 37034704 Free PMC article. Updated. Preprint.
References
-
- Xie, S., Duan, J., Li, B., Zhou, P. & Hon, G. C. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell66, 285–299.e5 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA1157910
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

