Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis
- PMID: 20631810
- PMCID: PMC3105664
- DOI: 10.1038/ismej.2010.88
Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis
Abstract
In patients afflicted with cystic fibrosis (CF), morbidity and mortality are primarily associated with the adverse consequences of chronic microbial bronchial infections, which are thought to be caused by a few opportunistic pathogens. However, recent evidence suggests the presence of other microorganisms, which may significantly affect the course and outcome of the infection. Using a combination of 16S rRNA gene clone libraries, bacterial culturing and pyrosequencing of barcoded 16S rRNA amplicons, the microbial communities present in CF patient sputum samples were examined. In addition to previously recognized CF pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus, >60 phylogenetically diverse bacterial genera that are not typically associated with CF pathogenesis were also detected. A surprisingly large number of fermenting facultative and obligate anaerobes from multiple bacterial phyla was present in each sample. Many of the bacteria and sequences found were normal residents of the oropharyngeal microflora and with many containing opportunistic pathogens. Our data suggest that these undersampled organisms within the CF lung are part of a much more complex microbial ecosystem than is normally presumed. Characterization of these communities is the first step in elucidating potential roles of diverse bacteria in disease progression and to ultimately facilitate advances in CF therapy.
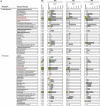
Figures


References
-
- Al Masalma M, Armougom F, Scheld WM, Dufour H, Roche PH, Drancourt M, et al. The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin Infect Dis. 2009;48:1169–1178. - PubMed
-
- Armougom F, Bittar F, Stremler N, Rolain JM, Robert C, Dubus JC, et al. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. Eur J Clin Microbiol Infect Dis. 2009;28:1151–1154. - PubMed
-
- Balough K, McCubbin M, Weinberger M, Smits W, Ahrens R, Fick R. The relationship between infection and inflammation in the early stages of lung disease from cystic fibrosis. Pediatr Pulmonol. 1995;20:63–70. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

