Artificial selection for determinate growth habit in soybean
- PMID: 20421496
- PMCID: PMC2889302
- DOI: 10.1073/pnas.1000088107
Artificial selection for determinate growth habit in soybean
Abstract
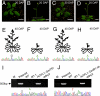
Determinacy is an agronomically important trait associated with the domestication in soybean (Glycine max). Most soybean cultivars are classifiable into indeterminate and determinate growth habit, whereas Glycine soja, the wild progenitor of soybean, is indeterminate. Indeterminate (Dt1/Dt1) and determinate (dt1/dt1) genotypes, when mated, produce progeny that segregate in a monogenic pattern. Here, we show evidence that Dt1 is a homolog (designated as GmTfl1) of Arabidopsis terminal flower 1 (TFL1), a regulatory gene encoding a signaling protein of shoot meristems. The transition from indeterminate to determinate phenotypes in soybean is associated with independent human selections of four distinct single-nucleotide substitutions in the GmTfl1 gene, each of which led to a single amino acid change. Genetic diversity of a minicore collection of Chinese soybean landraces assessed by simple sequence repeat (SSR) markers and allelic variation at the GmTfl1 locus suggest that human selection for determinacy took place at early stages of landrace radiation. The GmTfl1 allele introduced into a determinate-type (tfl1/tfl1) Arabidopsis mutants fully restored the wild-type (TFL1/TFL1) phenotype, but the Gmtfl1 allele in tfl1/tfl1 mutants did not result in apparent phenotypic change. These observations indicate that GmTfl1 complements the functions of TFL1 in Arabidopsis. However, the GmTfl1 homeolog, despite its more recent divergence from GmTfl1 than from Arabidopsis TFL1, appears to be sub- or neo-functionalized, as revealed by the differential expression of the two genes at multiple plant developmental stages and by allelic analysis at both loci.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Carter T, Nelson R, Sneller C, Cui Z. Soybeans: Improvement Production and Uses. Madison, WI: Am Soc of America, Crop Sci Soc of America, Soil Sci Soc of America; 2004. pp. 303–416.
-
- Li Y, et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet. 2008;117:857–871. - PubMed
-
- Ting CL. Genetic studies on the wild and cultivated soybeans. J Am Soc Agron. 1946;38:381–398.
-
- Nagata T. Studies on the Characteristics of Soybean Varieties. Tokyo: Jap Soybean Assoc; 1950. p. 115.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

