Presence of novel, potentially homoacetogenic bacteria in the rumen as determined by analysis of formyltetrahydrofolate synthetase sequences from ruminants
- PMID: 20118378
- PMCID: PMC2849231
- DOI: 10.1128/AEM.02580-09
Presence of novel, potentially homoacetogenic bacteria in the rumen as determined by analysis of formyltetrahydrofolate synthetase sequences from ruminants
Abstract
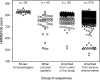
Homoacetogens produce acetate from H(2) and CO(2) via the Wood-Ljungdahl pathway. Some homoacetogens have been isolated from the rumen, but these organisms are expected to be only part of the full diversity present. To survey the presence of rumen homoacetogens, we analyzed sequences of formyltetrahydrofolate synthetase (FTHFS), a key enzyme of the Wood-Ljungdahl pathway. A total of 275 partial sequences of genes encoding FTHFS were PCR amplified from rumen contents of a cow, two sheep, and a deer. Phylogenetic trees were constructed using these FTHFS gene sequences and the translated amino acid sequences, together with other sequences from public databases and from novel nonhomoacetogenic bacteria isolated from the rumen. Over 90% of the FTHFS sequences fell into 34 clusters defined with good bootstrap support. Few rumen-derived FTHFS sequences clustered with sequences of known homoacetogens. Conserved residues were identified in the deduced FTHFS amino acid sequences from known homoacetogens, and their presence in the other sequences was used to determine a "homoacetogen similarity" (HS) score. A homoacetogen FTHFS profile hidden Markov model (HoF-HMM) was used to assess the homology of rumen and homoacetogen FTHFS sequences. Many clusters had low HS scores and HoF-HMM matches, raising doubts about whether the sequences originated from homoacetogens. In keeping with these findings, FTHFS sequences from nonhomoacetogenic bacterial isolates grouped in these clusters with low scores. However, sequences that formed 10 clusters containing no known isolates but representing 15% of our FTHFS sequences from rumen samples had high HS scores and HoF-HMM matches and so could represent novel homoacetogens.
Figures


References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Balch, W. E., S. Schobert, R. S. Tanner, and R. S. Wolfe. 1977. Acetobacterium, a new genus of hydrogen-oxidizing, carbon dioxide-reducing, anaerobic bacteria. Int. J. Syst. Bacteriol. 27:355-361.
-
- Bomar, M., H. Hippe, and B. Schink. 1991. Lithotrophic growth and hydrogen metabolism by Clostridium magnum. FEMS Microbiol. Lett. 67:347-349. - PubMed
-
- Braun, M., S. Schoberth, and G. Gottschalk. 1979. Enumeration of bacteria forming acetate from H2 and CO2 in anaerobic habitats. Arch. Microbiol. 120:201-204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

