Microorganisms with novel dissimilatory (bi)sulfite reductase genes are widespread and part of the core microbiota in low-sulfate peatlands
- PMID: 21169452
- PMCID: PMC3067207
- DOI: 10.1128/AEM.01352-10
Microorganisms with novel dissimilatory (bi)sulfite reductase genes are widespread and part of the core microbiota in low-sulfate peatlands
Abstract
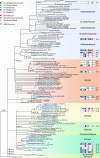
Peatlands of the Lehstenbach catchment (Germany) house as-yet-unidentified microorganisms with phylogenetically novel variants of the dissimilatory (bi)sulfite reductase genes dsrAB. These genes are characteristic of microorganisms that reduce sulfate, sulfite, or some organosulfonates for energy conservation but can also be present in anaerobic syntrophs. However, nothing is currently known regarding the abundance, community dynamics, and biogeography of these dsrAB-carrying microorganisms in peatlands. To tackle these issues, soils from a Lehstenbach catchment site (Schlöppnerbrunnen II fen) from different depths were sampled at three time points over a 6-year period to analyze the diversity and distribution of dsrAB-containing microorganisms by a newly developed functional gene microarray and quantitative PCR assays. Members of novel, uncultivated dsrAB lineages (approximately representing species-level groups) (i) dominated a temporally stable but spatially structured dsrAB community and (ii) represented "core" members (up to 1% to 1.7% relative abundance) of the autochthonous microbial community in this fen. In addition, denaturing gradient gel electrophoresis (DGGE)- and clone library-based comparisons of the dsrAB diversity in soils from a wet meadow, three bogs, and five fens of various geographic locations (distance of ∼1 to 400 km) identified that one Syntrophobacter-related and nine novel dsrAB lineages are widespread in low-sulfate peatlands. Signatures of biogeography in dsrB-based DGGE data were not correlated with geographic distance but could be explained largely by soil pH and wetland type, implying that the distribution of dsrAB-carrying microorganisms in wetlands on the scale of a few hundred kilometers is not limited by dispersal but determined by local environmental conditions.
Figures



References
-
- Alewell, C., B. Manderscheid, H. Meesenburg, and J. Bittersohl. 2000. Is acidification still an ecological threat? Nature 407:856-858. - PubMed
-
- Alewell, C., and M. Novak. 2001. Spotting zones of dissimilatory sulfate reduction in a forested catchment: the 34S-35S approach. Environ. Pollut. 112:369-377. - PubMed
-
- Alewell, C., S. Paul, G. Lischeid, K. Küsel, and M. Gehre. 2006. Characterizing the redox status in three different forested wetlands with geochemical data. Environ. Sci. Technol. 40:7609-7615. - PubMed
-
- Alewell, C., S. Paul, G. Lischeid, and F. R. Storck. 2008. Co-regulation of redox processes in freshwater wetlands as a function of organic matter availability? Sci. Total Environ. 404:335-342. - PubMed
-
- Blodau, C., B. Mayer, S. Peiffer, and T. R. Moore. 2007. Support for an anaerobic sulfur cycle in two Canadian peatland soils. J. Geophys. Res. 112:G02004.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

