Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies
- PMID: 21156758
- PMCID: PMC3025733
- DOI: 10.1093/aob/mcq235
Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies
Abstract
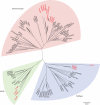
Background and aims: The genus Fritillaria (Liliaceae) comprises species with extremely large genomes (1C = 30 000-127 000 Mb) and a bicontinental distribution. Most North American species (subgenus Liliorhiza) differ from Eurasian Fritillaria species by their distinct phylogenetic position and increased amounts of heterochromatin. This study examined the contribution of major repetitive elements to the genome obesity found in Fritillaria and identified repeats contributing to the heterochromatin arrays in Liliorhiza species.
Methods: Two Fritillaria species of similar genome size were selected for detailed analysis, one from each phylogeographical clade: F. affinis (1C = 45·6 pg, North America) and F. imperialis (1C = 43·0 pg, Eurasia). Fosmid libraries were constructed from their genomic DNAs and used for identification, sequence characterization, quantification and chromosome localization of clones containing highly repeated sequences.
Key results and conclusions: Repeats corresponding to 6·7 and 4·7 % of the F. affinis and F. imperialis genome, respectively, were identified. Chromoviruses and the Tat lineage of Ty3/gypsy group long terminal repeat retrotransposons were identified as the predominant components of the highly repeated fractions in the F. affinis and F. imperialis genomes, respectively. In addition, a heterogeneous, extremely AT-rich satellite repeat was isolated from F. affinis. The FriSAT1 repeat localized in heterochromatic bands makes up approx. 26 % of the F. affinis genome and substantial genomic fractions in several other Liliorhiza species. However, no evidence of a relationship between heterochromatin content and genome size variation was observed. Also, this study was unable to reveal any predominant repeats which tracked the increasing/decreasing trends of genome size evolution in Fritillaria. Instead, the giant Fritillaria genomes seem to be composed of many diversified families of transposable elements. We hypothesize that the genome obesity may be partly determined by the failure of removal mechanisms to counterbalance effectively the retrotransposon amplification.
Figures



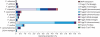
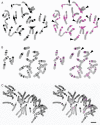

References
-
- Bakhshi Khaniki G. Giemsa C-banding and karyological studies in species of Rhinopetalum (Liliaceae) National Academy of Science Letters. 2004;27:399–411.
-
- Bennett MD, Leitch IJ. Plant DNA C-values database. 2005 (release 4·0, October 2005). http://data.kew.org/cvalues/ - PubMed
-
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London. B, Biological Sciences. 1976;274:227–274. - PubMed
-
- Darlington CD. The internal mechanics of the chromosomes I.-The nuclear cycle in Fritillaria. Proceedings of the Royal Society of London-Series B. 1935;118:33–59.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

