Genetic characterization of human T-cell lymphotropic virus type 1 in Mozambique: transcontinental lineages drive the HTLV-1 endemic
- PMID: 21532745
- PMCID: PMC3075232
- DOI: 10.1371/journal.pntd.0001038
Genetic characterization of human T-cell lymphotropic virus type 1 in Mozambique: transcontinental lineages drive the HTLV-1 endemic
Abstract
Background: Human T-Cell Lymphotropic Virus Type 1 (HTLV-1) is the etiological agent of adult T-cell leukemia (ATL) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). It has been estimated that 10-20 million people are infected worldwide, but no successful treatment is available. Recently, the epidemiology of this virus was addressed in blood donors from Maputo, showing rates from 0.9 to 1.2%. However, the origin and impact of HTLV endemic in this population is unknown.
Objective: To assess the HTLV-1 molecular epidemiology in Mozambique and to investigate their relationship with HTLV-1 lineages circulating worldwide.
Methods: Blood donors and HIV patients were screened for HTLV antibodies by using enzyme immunoassay, followed by Western Blot. PCR and sequencing of HTLV-1 LTR region were applied and genetic HTLV-1 subtypes were assigned by the neighbor-joining method. The mean genetic distance of Mozambican HTLV-1 lineages among the genetic clusters were determined. Human mitochondrial (mt) DNA analysis was performed and individuals classified in mtDNA haplogroups.
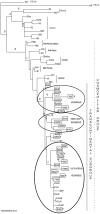
Results: LTR HTLV-1 analysis demonstrated that all isolates belong to the Transcontinental subgroup of the Cosmopolitan subtype. Mozambican HTLV-1 sequences had a high inter-strain genetic distance, reflecting in three major clusters. One cluster is associated with the South Africa sequences, one is related with Middle East and India strains and the third is a specific Mozambican cluster. Interestingly, 83.3% of HIV/HTLV-1 co-infection was observed in the Mozambican cluster. The human mtDNA haplotypes revealed that all belong to the African macrohaplogroup L with frequencies representatives of the country.
Conclusions: The Mozambican HTLV-1 genetic diversity detected in this study reveals that although the strains belong to the most prevalent and worldwide distributed Transcontinental subgroup of the Cosmopolitan subtype, there is a high HTLV diversity that could be correlated with at least 3 different HTLV-1 introductions in the country. The significant rate of HTLV-1a/HIV-1C co-infection, particularly in the Mozambican cluster, has important implications for the controls programs of both viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kalyanaraman VS, Sarngadharan MG, Robert-Guroff M, Miyoshi I, Golde D, et al. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. - PubMed
-
- Mahieux R, Ibrahim F, Mauclere P, Herve V, Michel P, et al. Molecular epidemiology of 58 new African human T-cell leukemia virus type 1 (HTLV-1) strains: Identification of a new and distinct HTLV-1 molecular subtype in central Africa and in pygmies. Journal of Virology. 1997;71:1317–1333. - PMC - PubMed
-
- Salemi M, Desmyter J, Vandamme AM. Tempo and mode of human and simian T-lymphotropic virus (HTLV/STLV) evolution revealed by analyses of full-genome sequences. Mol Biol Evol. 2000;17:374–386. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

