Old gene duplication facilitates origin and diversification of an innovative communication system--twice
- PMID: 21127261
- PMCID: PMC3009798
- DOI: 10.1073/pnas.1011803107
Old gene duplication facilitates origin and diversification of an innovative communication system--twice
Abstract
The genetic basis of parallel innovation remains poorly understood due to the rarity of independent origins of the same complex trait among model organisms. We focus on two groups of teleost fishes that independently gained myogenic electric organs underlying electrical communication. Earlier work suggested that a voltage-gated sodium channel gene (Scn4aa), which arose by whole-genome duplication, was neofunctionalized for expression in electric organ and subsequently experienced strong positive selection. However, it was not possible to determine if these changes were temporally linked to the independent origins of myogenic electric organs in both lineages. Here, we test predictions of such a relationship. We show that Scn4aa co-option and rapid sequence evolution were tightly coupled to the two origins of electric organ, providing strong evidence that Scn4aa contributed to parallel innovations underlying the evolutionary diversification of each electric fish group. Independent evolution of electric organs and Scn4aa co-option occurred more than 100 million years following the origin of Scn4aa by duplication. During subsequent diversification of the electrical communication channels, amino acid substitutions in both groups occurred in the same regions of the sodium channel that likely contribute to electric signal variation. Thus, the phenotypic similarities between independent electric fish groups are also associated with striking parallelism at genetic and molecular levels. Our results show that gene duplication can contribute to remarkably similar innovations in repeatable ways even after long waiting periods between gene duplication and the origins of novelty.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
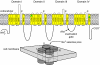
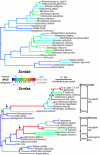

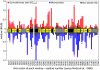

Comment in
-
How an ancient genome duplication electrified modern fish.Proc Natl Acad Sci U S A. 2010 Dec 21;107(51):21953-4. doi: 10.1073/pnas.1016298108. Epub 2010 Dec 13. Proc Natl Acad Sci U S A. 2010. PMID: 21149707 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

