Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence
- PMID: 21282452
- PMCID: PMC3067157
- DOI: 10.1128/AAC.01275-10
Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence
Abstract
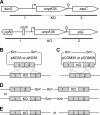
OmpK35 and OmpK36 are the major outer membrane porins of Klebsiella pneumoniae. In this study, a virulent clinical isolate was selected to study the role of these two porins in antimicrobial resistance and virulence. The single deletion of ompK36 (ΔompK36) resulted in MIC shifts of cefazolin, cephalothin, and cefoxitin from susceptible to resistant, while the single deletion of ompK35 (ΔompK35) had no significant effect. A double deletion of ompK35 and ompK36 (ΔompK35/36) further increased these MICs to high-level resistance and led to 8- and 16-fold increases in the MICs of meropenem and cefepime, respectively. In contrast to the routine testing medium, which is of high osmolarity, susceptibility tests using low-osmolarity medium showed that the ΔompK35 mutation resulted in a significant (≥ 4-fold) increase in the MICs of cefazolin and ceftazidime, whereas a ΔompK36 deletion conferred a significantly (4-fold) lower increase in the MIC of cefazolin. In the virulence assays, a significant (P < 0.05) defect in the growth rate was found only in the ΔompK35/36 mutant, indicating the effect on metabolic fitness. A significant (P < 0.05) increase in susceptibility to neutrophil phagocytosis was observed in both ΔompK36 and ΔompK35/36 mutants. In a mouse peritonitis model, the ΔompK35 mutant showed no change in virulence, and the ΔompK36 mutant exhibited significantly (P < 0.01) lower virulence, whereas the ΔompK35/36 mutant presented the highest 50% lethal dose of these strains. In conclusion, porin deficiency in K. pneumoniae could increase antimicrobial resistance but decrease virulence at the same time.
Figures

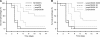
References
-
- Achouak, W., T. Heulin, and J.-M. Pagès. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199:1-7. - PubMed
-
- Andersen, J., S. A. Forst, K. Zhao, M. Inouye, and N. Delihas. 1989. The function of micF RNA: micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961-17970. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

