Genomic and functional analyses of Rhodococcus equi phages ReqiPepy6, ReqiPoco6, ReqiPine5, and ReqiDocB7
- PMID: 21097585
- PMCID: PMC3020559
- DOI: 10.1128/AEM.01952-10
Genomic and functional analyses of Rhodococcus equi phages ReqiPepy6, ReqiPoco6, ReqiPine5, and ReqiDocB7
Abstract
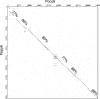
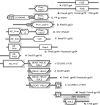
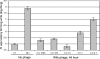
The isolation and results of genomic and functional analyses of Rhodococcus equi phages ReqiPepy6, ReqiDocB7, ReqiPine5, and ReqiPoco6 (hereafter referred to as Pepy6, DocB7, Pine5, and Poco6, respectively) are reported. Two phages, Pepy6 and Poco6, more than 75% identical, exhibited genome organization and protein sequence likeness to Lactococcus lactis phage 1706 and clostridial prophage elements. An unusually high fraction, 27%, of Pepy6 and Poco6 proteins were predicted to possess at least one transmembrane domain, a value much higher than the average of 8.5% transmembrane domain-containing proteins determined from a data set of 36,324 phage protein entries. Genome organization and protein sequence comparisons place phage Pine5 as the first nonmycobacteriophage member of the large Rosebush cluster. DocB7, which had the broadest host range among the four isolates, was not closely related to any phage or prophage in the database, and only 23 of 105 predicted encoded proteins could be assigned a functional annotation. Because of the relationship of Rhodococcus to Mycobacterium, it was anticipated that these phages should exhibit some of the features characteristic of mycobacteriophages. Traits that were identified as shared by the Rhodococcus phages and mycobacteriophages include the prevalent long-tailed morphology and the presence of genes encoding LysB-like mycolate-hydrolyzing lysis proteins. Application of DocB7 lysates to soils amended with a host strain of R. equi reduced recoverable bacterial CFU, suggesting that phage may be useful in limiting R. equi load in the environment while foals are susceptible to infection.
Figures




References
-
- Ackermann, H. W. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227-243. - PubMed
-
- Adindla, S., and L. Guruprasad. 2003. Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. J. Biosci. 28:169-179. - PubMed
-
- Akoh, C. C., G. C. Lee, Y. C. Liaw, T. H. Huang, and J. F. Shaw. 2004. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43:534-552. - PubMed
-
- Alvarez, V. M., et al. 2008. Bioremediation potential of a tropical soil contaminated with a mixture of crude oil and production water. J. Microbiol. Biotechnol. 18:1966-1974. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

