Wild chimpanzees infected with 5 Plasmodium species
- PMID: 21122230
- PMCID: PMC3294549
- DOI: 10.3201/eid1612.100424
Wild chimpanzees infected with 5 Plasmodium species
Abstract
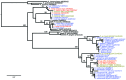
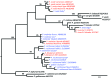
Data are missing on the diversity of Plasmodium spp. infecting apes that live in their natural habitat, with limited possibility of human-mosquito-ape exchange. We surveyed Plasmodium spp. diversity in wild chimpanzees living in an undisturbed tropical rainforest habitat and found 5 species: P. malariae, P. vivax, P. ovale, P. reichenowi, and P. gaboni.
Figures
References
-
- Coatney GR. The simian malarias: zoonoses, anthroponoses, or both? Am J Trop Med Hyg. 1971;20:795–803. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
