Validated and longitudinally stable asthma phenotypes based on cluster analysis of the ADEPT study
- PMID: 27978840
- PMCID: PMC5159977
- DOI: 10.1186/s12931-016-0482-9
Validated and longitudinally stable asthma phenotypes based on cluster analysis of the ADEPT study
Abstract
Background: Asthma is a disease of varying severity and differing disease mechanisms. To date, studies aimed at stratifying asthma into clinically useful phenotypes have produced a number of phenotypes that have yet to be assessed for stability and to be validated in independent cohorts. The aim of this study was to define and validate, for the first time ever, clinically driven asthma phenotypes using two independent, severe asthma cohorts: ADEPT and U-BIOPRED.
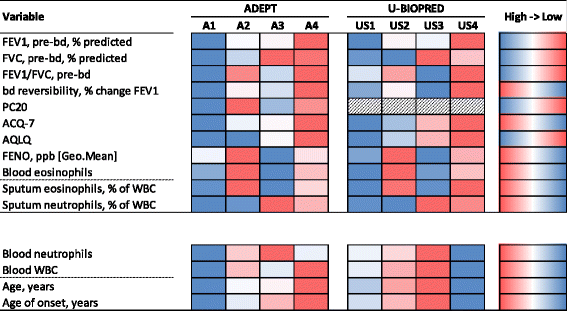
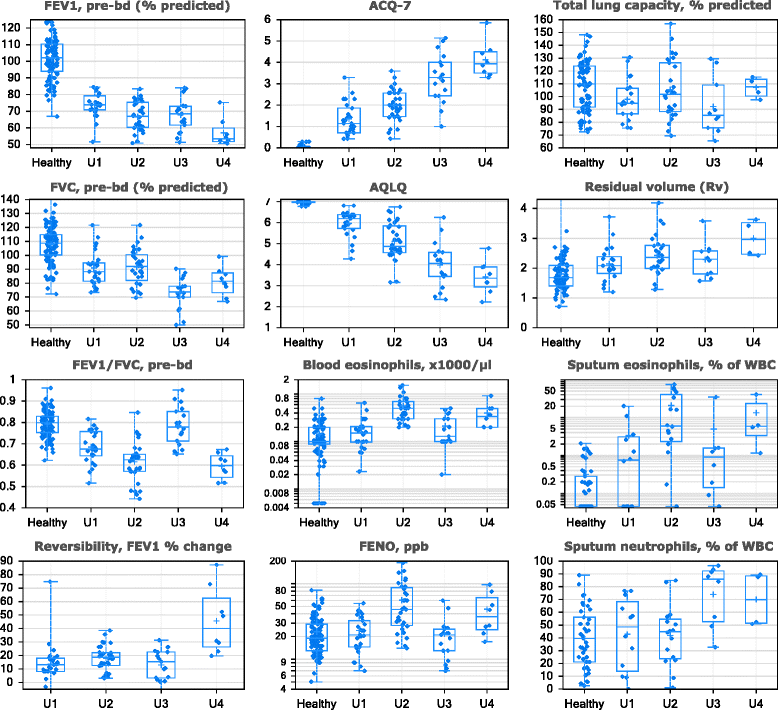
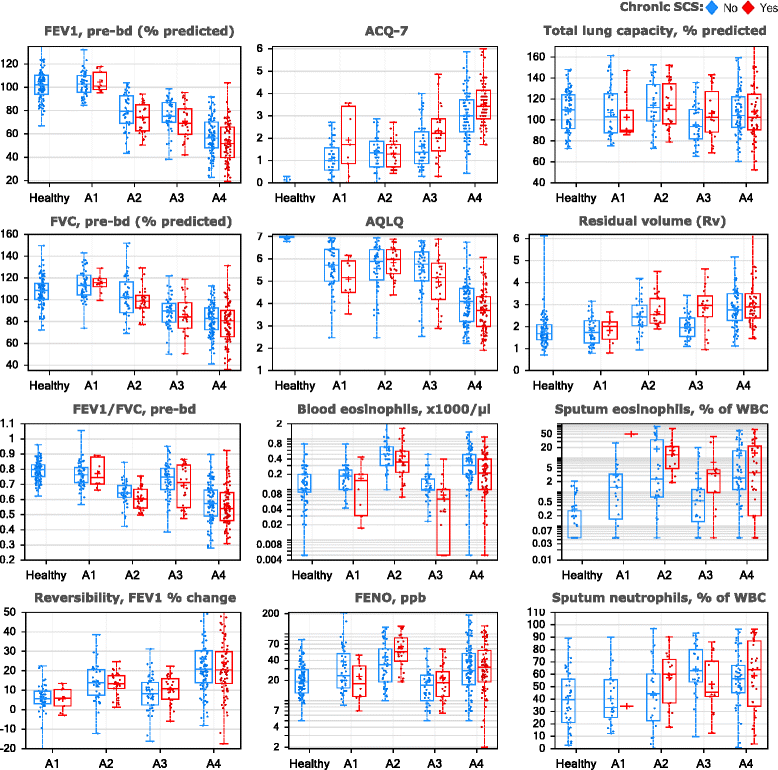
Methods: Fuzzy partition-around-medoid clustering was performed on pre-specified data from the ADEPT participants (n = 156) and independently on data from a subset of U-BIOPRED asthma participants (n = 82) for whom the same variables were available. Models for cluster classification probabilities were derived and applied to the 12-month longitudinal ADEPT data and to a larger subset of the U-BIOPRED asthma dataset (n = 397). High and low type-2 inflammation phenotypes were defined as high or low Th2 activity, indicated by endobronchial biopsies gene expression changes downstream of IL-4 or IL-13.
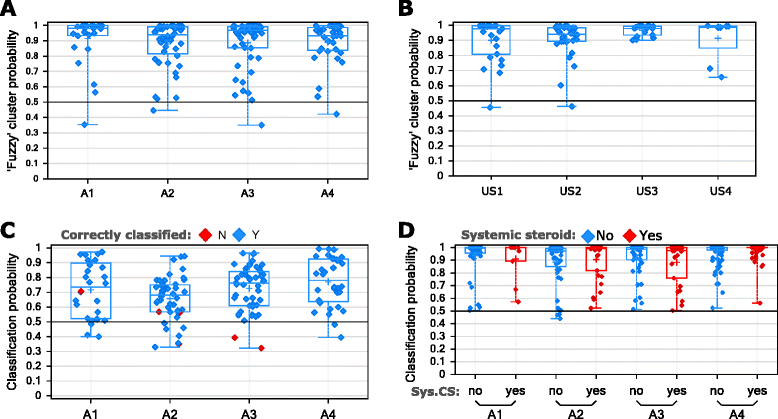
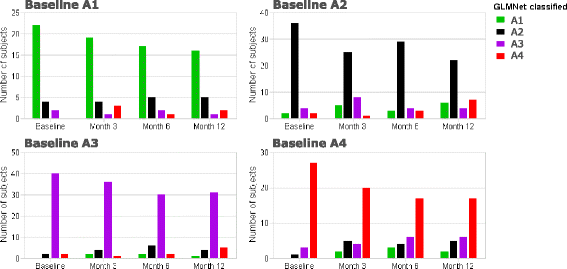
Results: Four phenotypes were identified in the ADEPT (training) cohort, with distinct clinical and biomarker profiles. Phenotype 1 was "mild, good lung function, early onset", with a low-inflammatory, predominantly Type-2, phenotype. Phenotype 2 had a "moderate, hyper-responsive, eosinophilic" phenotype, with moderate asthma control, mild airflow obstruction and predominant Type-2 inflammation. Phenotype 3 had a "mixed severity, predominantly fixed obstructive, non-eosinophilic and neutrophilic" phenotype, with moderate asthma control and low Type-2 inflammation. Phenotype 4 had a "severe uncontrolled, severe reversible obstruction, mixed granulocytic" phenotype, with moderate Type-2 inflammation. These phenotypes had good longitudinal stability in the ADEPT cohort. They were reproduced and demonstrated high classification probability in two subsets of the U-BIOPRED asthma cohort.
Conclusions: Focusing on the biology of the four clinical independently-validated easy-to-assess ADEPT asthma phenotypes will help understanding the unmet need and will aid in developing tailored therapies.
Trial registration: NCT01274507 (ADEPT), registered October 28, 2010 and NCT01982162 (U-BIOPRED), registered October 30, 2013.
Keywords: Biological markers; Cluster analysis; Observational study.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

